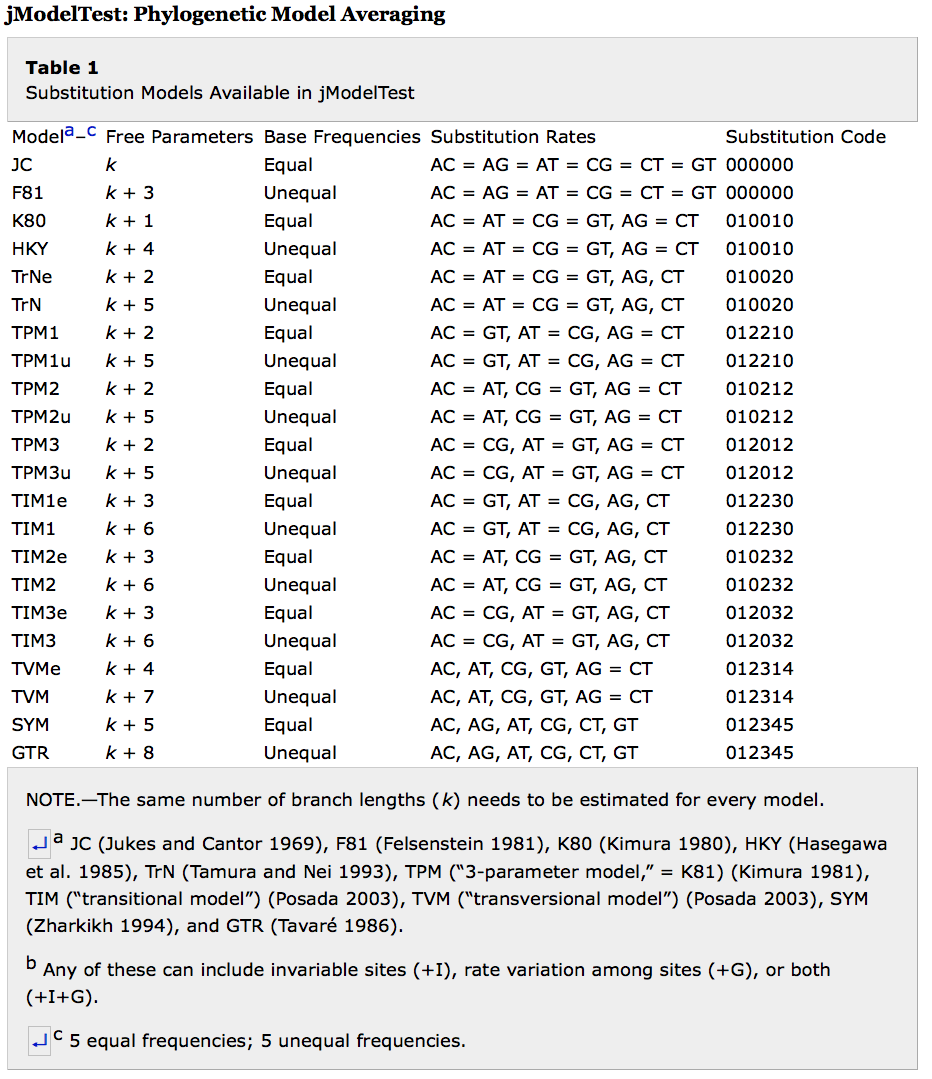
SSM (standard substitution models) is a BEAST 2 package containing the following standard time-reversible substitution models: JC, F81, K80, HKY, TrNf, TrN, TPM1, TPM1f, TPM2, TPM2f, TPM3, TPM3f, TIM1, TIM1f, TIM2, TIM2f, TIM3 , TIM3f, TVMf, TVM, SYM, GTR.
They are summarised in Table 1 in jModelTest (Posada 2008):
Rates are estimated assuming a Dirichlet(1,1,1,1,1,1) prior on the 6 rates (AC, AG, AT, CG, CT, GT). For models that use fewer parameters, e.g. HKY, all six associated rates are logged.
Models have a Dirichlet(1,1,1,1) prior on frequencies (in case they are estimated).
Remco Bouckaert, & Dong Xie. (2017, September 25). Standard Nucleotide Substitution Models v1.0.1. http://doi.org/10.5281/zenodo.995740
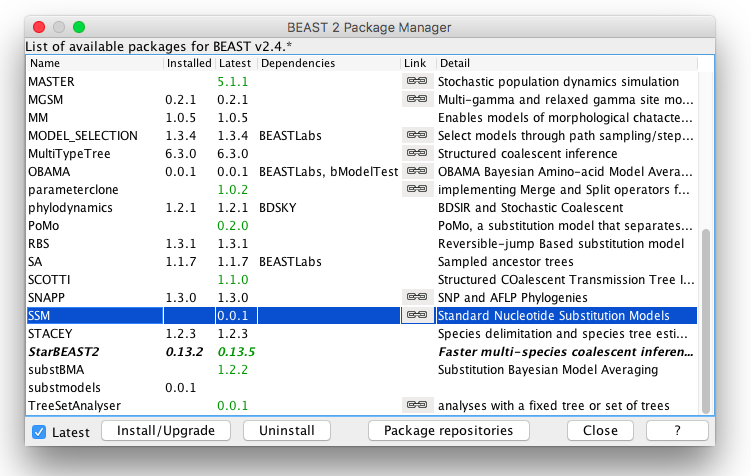
To install SSM, it is easiest to start BEAUti (a program that is part of BEAST), and select the menu File/Manage packages. A package manager dialog pops up, that looks something like this:
If the SSM package is listed, just click on it to select it, and hit the Install/Upgrade button.
If the SSM package is not listed, you may need to add a package repository by clicking the "Package repositories" button. A window pops up where you can click "Add URL" and add "https://raw.githubusercontent.com/CompEvol/CBAN/master/packages-extra.xml" in the entry. After clicking OK, the dialog should look something like this:
Click OK and now SSM should be listed in the package manager (as in the first dialog above). Select and click Install/Upgrade to install.
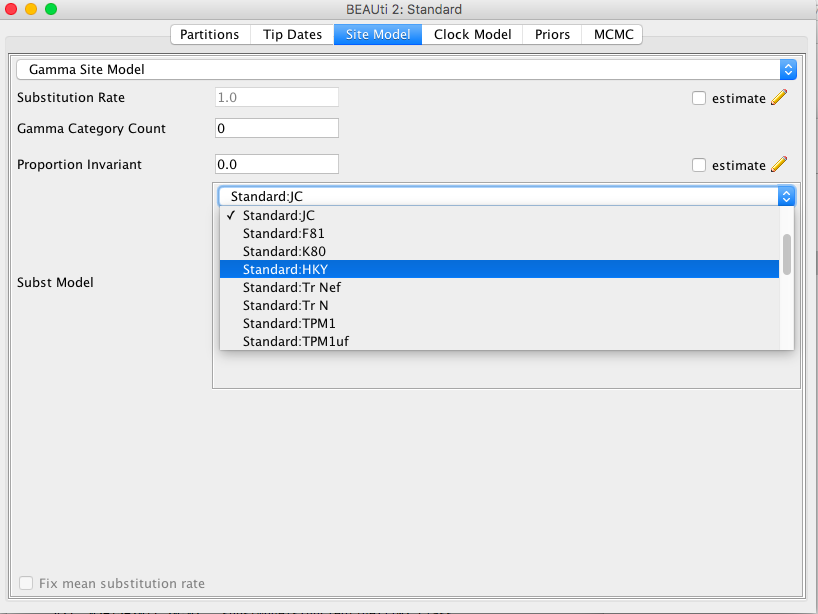
To use one of the standard models, import a nucleotide alignment in BEAUti, and select the site model pannel. Under the "substitution models" drop down box, you will find the models, just pick the one you want to use.