Introduction to Open Science¶
+Learning Objectives
+After this lesson, you should be able to:
+-
+
- Explain what Open Science is +
- Explain the components of Open Science +
- Describe the behaviors of Open Science +
- Explain why Open Science matters in education, research, and society +
- Understand the advantages and the challenges to Open Science +
+
What is Open Science?¶
+"Open Science is transparent and accessible knowledge that is shared and developed through collaborative networks"
+-Vincente-Saez & Martinez-Fuentes 2018
+
+
+
+
+
"Open Science is a collaborative and transparent approach to scientific research that emphasizes the accessibility, sharing, and reproducibility of data, methodologies, and findings to foster innovation and inclusivity"
+-ChatGPT
+
+
+
+
+
"A series of reforms that interrogate every step in the research life cycle to make it more efficient, powerful and accountable in our emerging digital society".
+-Jeffrey Gillan
+ +
+
+
+
+
Other Definitions
+"Open Science is defined as an inclusive construct that combines various movements and practices aiming to make multilingual scientific knowledge openly available, accessible and reusable for everyone, to increase scientific collaborations and sharing of information for the benefits of science and society, and to open the processes of scientific knowledge creation, evaluation and communication to societal actors beyond the traditional scientific community." - UNESCO Definition
+ +"Open Science is the movement to make scientific research (including publications, data, physical samples, and software) and its dissemination accessible to all levels of society, amateur or professional..." Wikipedia definition
+Foundational Open Science Skills¶
+1. Building a culture of scientists eager to share research materials - such as data, code, methods, documentation, and early results - with colleagues and society at large, in addition to traditional publications
+
+
2. Mastery of digital tools to create reproducible science that others can build upon
+
+
3. Understanding the push towards increased transparency and accountability for those practicing science (ie., compliance)
+
+
+
+
+
 +
+
+
+
+
+
What is Open Science | The Royal Society +
+
+
+
+
+
2023: the Year of Open Science¶
+The White House, joined by 10 federal agencies, and a coalition of more than 85 universities, declared 2023 the Year of Open Science as a way to bring awareness to the benefits of Open Science and to steer the scientitic community towards its adoption.
+NASA leads a prominent program called Transform to Open Science which includes an online class on Open Science.
+
+
+
+
+
+
6 Pillars of Open Science¶
+Open Access Publications
+Open Data
+Open Educational Resources
+Open Methodology
+Open Peer Review
+Open Source Software
+ +
+
+
+
+
Open Access Publications¶
+ +Definition
+"Open access is a publishing model for scholarly communication that makes research information available to readers at no cost, as opposed to the traditional subscription model in which readers have access to scholarly information by paying a subscription (usually via libraries)." -- OpenAccess.nl
+
+
+
Open Access Journal Examples
+Major publishers have provided access points for publishing your work
+ +
+
Types of Publishing Business Models:¶
+-
+
-
+
Subscription model - the author pays a smaller fee (or no fee) for the article to be published. The publisher then sells subscription access to the article (usually to institutes of higher education).
+
+ -
+
Open Access model - The author pays a larger fee to make the article freely available to anyone through a Creative Commons license.
+-
+
- Open Access publishing in Nature costs $12,290! +
- Open Access publising in PlosOne costs $2,290 +
+
+
+
Research Article Versions¶
+-
+
-
+
Preprint - In academic publishing, a preprint is a version of scholary paper that precedes formal peer-review and publication in a scientific journal. The preprint may be available, often as a non-typeset version available for free online.
+++Pre-print Services
+-
+
- ASAPbio Pre-Print Server List - ASAPbio is a scientist-driven non-profit promoting transparency and innovation comprehensive list of pre-print servers inthe field of life science communication. +
- ESSOar - Earth and Space Science Open Archive hosted by the American Geophysical Union. +
- Peer Community In (PCI) a free recommendation process of scientific preprints based on peer reviews +
- OSF.io Preprints are partnered with numerous projects under the "-rXivs" +
++The rXivs
+-
+
- + + +
- + + +
- + + +
-
+
arXiv - is a free distribution service and an open-access archive for 2,086,431 scholarly articles in the fields of physics, mathematics, computer science, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics.
+
+ - + + +
- BioRxiv - is an open access preprint repository for the biological sciences. +
- BodorXiv +
- EarthArXiv - is an open access preprint repository for the Earth sciences. +
- EcsArXiv - a free preprint service for electrochemistry and solid state science and technology +
- EdArXiv - for the education research community +
- EngrXiv for the engineering community +
- EvoEcoRxiv - is an open acccess preprint repository for Evolutionary and Ecological sciences. +
- MediArXiv for Media, Film, & Communication Studies +
- MedRxiv - is an open access preprint repository for Medical sciences. +
- PaleorXiv - is an open access preprint repository for Paleo Sciences +
- PsyrXiv - is an open access preprint repository for Psychological sciences. +
- SocArXiv - is an open access preprint repository for Social sciences. +
- SportrXiv - is an open access preprint for Sports sciences. +
- ThesisCommons - open Theses +
+ -
+
Author's accepted manuscript (AAM) - includes changes that came about during peer-review process. It is a non-typeset or formatted article. This often had an embargo period of 12-24 months
+
+ -
+
Published version of record (VOR) - includes stylistic edits, online & print formatting. This is the version that publishers claim ownership of with copyrights or exclusive licensing.
+
+
+
Copyrights and Science Publishing
+Upon completion of a peer-reviewed science paper, the author typically 1. signs over the copyright of the paper to the publisher or 2. signs an exclusive license agreement with the publisher
+For example authors that publish in Science retain their copyright but sign a 'license to pubish' agreement with AAAS
+Elsevier requires authors to sign over copyright of the article but authors retains some rights of distribution
+ +
+
New Open Access Mandates in US¶
+The White House Office of Science and Technology (OSTP) has recently released a policy document known as the Nelson Memo stating that tax-payer funded research must by open access by 2026 with no embargo period.
+Authors can comply with the memo by either:
+-
+
- Publishing Open Access (this usually requires higher fees) +
- Distributing the Author's Accepted Manuscript (AAM) +
Read NSF's open access plan in reponse to the Nelson Memo
+Read USDA's open access plan in reponse to the Nelson Memo
+
+
+
+
Additional Info¶
+University of Arizona Libraries information on Open Access publishing including agreements with several journals to reduce or waive publishing fees.
+ +
+
+
+
+
+
Open Data¶
+Definitions
+“Open data and content can be freely used, modified, and shared by anyone for any purpose” - The Open Definition
+"Open data is data that can be freely used, re-used and redistributed by anyone - subject only, at most, to the requirement to attribute and sharealike." - Open Data Handbook
+ +
+
+
Data are the foundation for any scientific endeavor. A lot of thought needs to go into how to best collect, store, analyze, curate, share, and archive data.
+
+
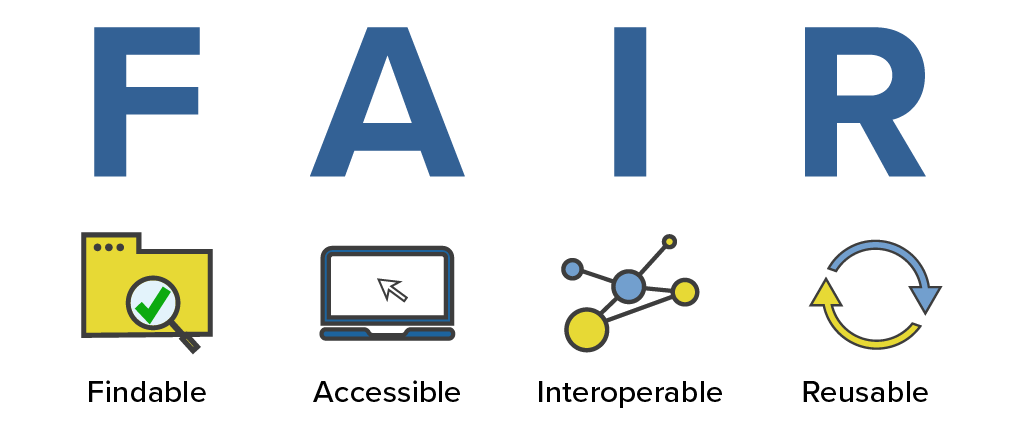
FAIR Principles¶
+In 2016, the FAIR Guiding Principles for scientific data management and stewardship were published in Scientific Data.
+Findable: +Making data discoverable by the wider academic community and the public
+Accessible: +Using unique identifiers, metadata and a clear use of language and access protocols
+Interoperable: +Applying standards to encode and exchange data and metadata
+Reusable: +Enabling the repurposing of researach outputs to maximize their research potential
+
+
+
+
Reasons to Make your Data Open
+-
+
- Unnecessary duplication. Duplication of research is costly for society, and places unnecessary burden on heavily researched people and populations. +
- The data underlying publications are maintained and accessible, allowing for validation of results. +
- Data openness leads to more collaboration and advances research and innovation. +
- Your research is more visible and has greater impact. Publications which allow access to the underlying data get more citations. Greater visibility also allows for better validation and scrutiny of findings. +
- Other researchers can cite your data, which will drive up your citation number and increase your influence in your field of research. +
- Storing your data in a public repository also provides you with secure and ongoing storage that may otherwise not be available to you. +-Foster Open Science +
+
+
+
As Open as Possible, as Closed as Necessary¶
+There are many circumstances where open data could be harmful:
+-
+
-
+
Data on human health
+
+ -
+
Location of endangered species or archaeological sites
+
+ -
+
Data that individuals or groups do not want to be public
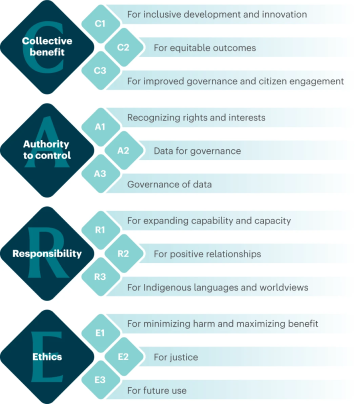
+++CARE Principles
+The CARE Principles for Indigenous Data Governance were drafted at the International Data Week and Research Data Alliance Plenary co-hosted event "Indigenous Data Sovereignty Principles for the Governance of Indigenous Data Workshop," 8 November 2018, Gaborone, Botswana.
+Collective Benefit
+-
+
- C1. For inclusive development and innovation +
- C2. For improved governance and citizen engagement +
- C3. For equitable outcomes +
Authority to Control
+-
+
- A1. Recognizing rights and interests +
- A2. Data for governance +
- A3. Governance of data +
Responsibility
+-
+
- R1. For positive relationships +
- R2. For expanding capability and capacity +
- R3. For Indigenous languages and worldviews +
Ethics
+-
+
- E1. For minimizing harm and maximizing benefit +
- E2. For justice +
- E3. For future use +
+ -
+
Data for making lethal weapons
+
+
+
Open vs. FAIR
+FAIR does not demand that data be open: See one definition of open: http://opendefinition.org/
+Open data does not necessarily mean it is FAIR
+
+
Additional Info¶
+-
+
-
+
The Ethics of Geolocated Data from UK Statistics Authority
+
+ -
+
Health information US HIPAA
+
+ -
+
Indigenous data sovereignty: CARE Principles for Indigenous Data Governance , Global Indigenous Data Alliance (GIDA), First Nations OCAP® (Ownership Control Access and Possession), Circumpolar Inuit Protocols for Equitable and Ethical Engagement
+
+
+
+
+
+
+
+
Open Educational Resources¶
+Definitions
+"Open Educational Resources (OER) are learning, teaching and research materials in any format and medium that reside in the public domain or are under copyright that have been released under an open license, that permit no-cost access, re-use, re-purpose, adaptation and redistribution by others." - UNESCO
+ +Digital Literacy Organizations
+-
+
- The Carpentries - teaches foundational coding and data science skills to researchers worldwide +
- EdX - Massively Open Online Courses (not all open) hosted through University of California Berkeley +
- EveryoneOn - mission is to unlock opportunity by connecting families in underserved communities to affordable internet service and computers, and delivering digital skills trainings +
- ConnectHomeUSA - is a movement to bridge the digital divide for HUD-assisted housing residents in the United States under the leadership of national nonprofit EveryoneOn +
- Global Digital Literacy Council - has dedicated more than 15 years of hard work to the creation and maintenance of worldwide standards in digital literacy +
- IndigiData - training and engaging tribal undergraduate and graduate students in informatics +
- National Digital Equity Center a 501c3 non-profit, is a nationally recognized organization with a mission to close the digital divide across the United States +
- National Digital Inclusion Allaince - advances digital equity by supporting community programs and equipping policymakers to act +
- Net Literacy +
- Open Educational Resources Commons +
- Project Pythia is the education working group for Pangeo and is an educational resource for the entire geoscience community +
- Research Bazaar - is a worldwide festival promoting the digital literacy emerging at the centre of modern research +
- TechBoomers - is an education and discovery website that provides free tutorials of popular websites and Internet-based services in a manner that is accessible to older adults and other digital technology newcomers +
Educational Materials
+-
+
- Teach Together by Greg Wilson +
- DigitalLearn +
+
+
+
+
+
+
Open Methodology¶
+Definitions
+"An open methodology is simply one which has been described in sufficient detail to allow other researchers to repeat the work and apply it elsewhere." - Watson (2015)
+"Open Methodology refers to opening up methods that are used by researchers to achieve scientific results and making them publicly available." - Open Science Network Austria
+
+
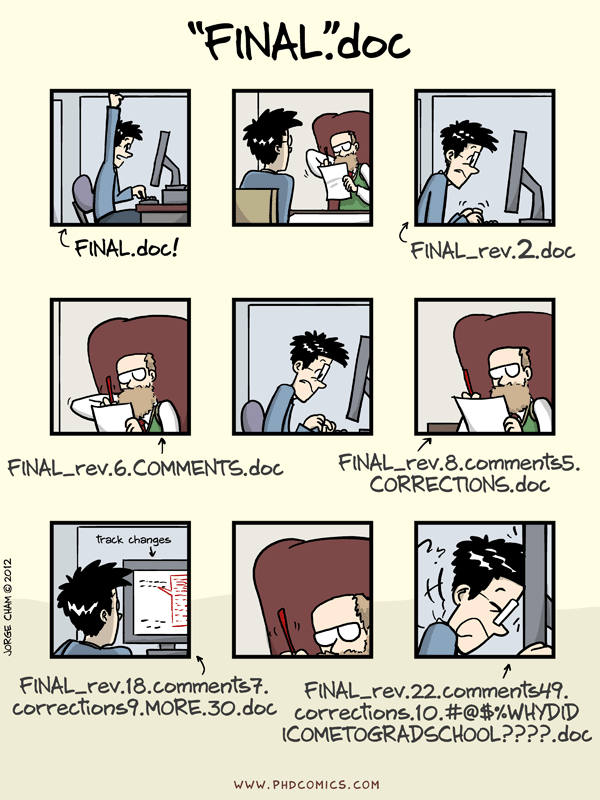
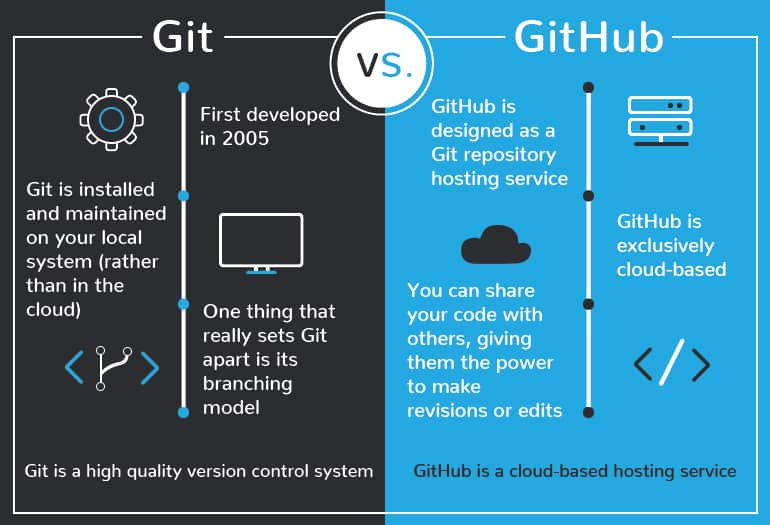
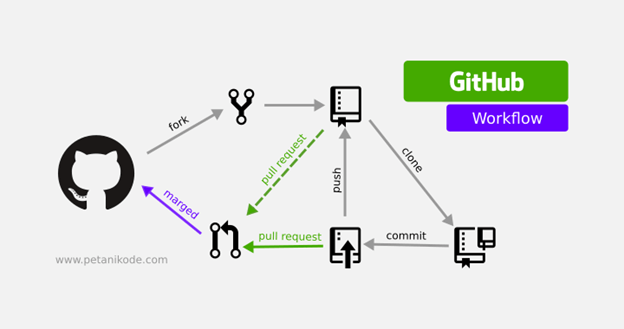
Sharing Research Computer Code¶
+Scientists around the globe are creating computer code for scientific analysis. These are valuable contributions that need to be shared!
+Platforms like GitHub and GitLab are ideal for collaboratively developing code and sharing with the open internet. In FOSS, we will show you how to use Github for sharing code, documentation, hosting websites, and software version control.
+ +
+  +
+
+
+
+
Publishing Your Methods or Protocols¶
+Platforms for Publishing Protocols & Bench Techniques
+-
+
- BioProtocol +
- Current Protocols +
- Gold Biotechnology Protocol list +
- JoVE - Journal of Visualized Experiments +
- Nature Protocols +
- OpenWetWare +
- Protocol Exchange +
- Protocols Online +
- Protocols +
- SciGene +
- Springer Nature Experiments +
+
+
+
PreRegistration¶
+Preregistration is detailing your research and analysis plan and submitting it to an online registry before you engage in the research.
+Why Do This?¶
+Preregistration makes your process more open and records the difference between your initial research plan what you end up actually doing.
+Preregistration separates hypothesis-generating (exploratory) from hypothesis-testing (confirmatory) research. Both are important. But the same data cannot be used to generate and test a hypothesis, which can happen unintentionally and reduce the credibility of your results.
+It also helps us avoid practices like p-hacking or Hypothesizing After the Results are Known(HARKing).
+Additional Info¶
+Read this publication by Nosek et al. 2018
+Open Science Framework Preregistration https://www.cos.io/initiatives/prereg
+
+
+
+
+
+
+
Open Peer Review¶
+
+
Definitions
+Open peer review is an umbrella term for a number of overlapping ways that peer review models can be adapted in line with the aims of Open Science, including making reviewer and author identities open, publishing review reports and enabling greater participation in the peer review process.
+ +
+
+
Traditional Closed Peer-Review System¶
+-
+
- Throughout and after the process, the author remains unaware of the reviewers' identities, while the reviewers know the identity of the authors. +
- All communications between authors, reviewers and editors remains private +
+
+
Complaints with the Traditional Closed Peer-Review System¶
+-
+
- Unreliable and Inconsistent +
- Delays and Expense +
- Lack of Accountability and Risks of Subversion +
- Social and Publication Biases +
- Lack of Incentives +
+
+
+
Open Peer-Review Ideas¶
+ +
+
+
+
Defenders of the Traditional Peer-Review System
+
+
+
Example Open Peer-Review Systems
+F1000Research An open research publishing platform that offers open peer review and rapid publication. +The article from Ross-Hellauer et al. (2017) has open peer-reviews.
+
+
+
+
+
+
+
Open Source Software¶
+ +Definitions
+"Open source software is code that is designed to be publicly accessible—anyone can see, modify, and distribute the code as they see fit. Open source software is developed in a decentralized and collaborative way, relying on peer review and community production." - Red Hat
+ +
+
Research science (and also many companies) rely on open source software to operate
+
+
Open Source Software
+-
+
- Linux operating system and shell +
- Python +
- R +
- git +
- Conda +
- Docker +
- Cyverse +
- Pytorch +
- Tyson's Awesome List +
+
When you create a new software, library, or package, you become its parent and guardian.
+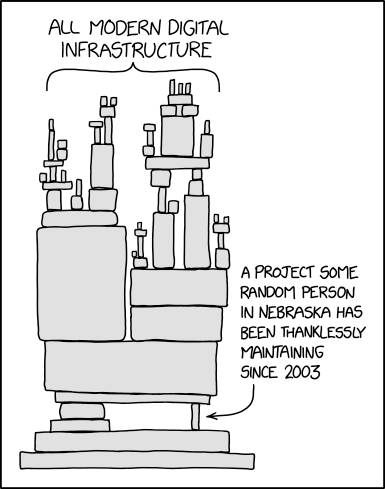 +
+
+
+
+
+
+
+
WHY do Open Science?¶
+A paper from Bartling & Friesike (2014) posits that there are 5 main schools of thought in Open Science, which represent 5 underlying motivations:
+-
+
- Democratic school: primarily concerned with making scholarly work freely available to everyone +
- Pragmatic school: primarily concerned with improving the quality of scholarly work by fostering collaboration and improving critiques +
- Infrastructure school: primarily focused on the platforms, tools, and services necessary to conduct efficient research, collaboration, and communication +
- Public school: primarily concerned with societal impact of scholarly work, focusing on engagement with broader public via citizen science, understandable scientific communication, and less formal communication +
- Measurement school: primarily concerned with the existing focus on journal publications as a means of measuring scholarly output, and focused on developing alternative measurements of scientific impact +
 +
+
We have added another school of thought
+-
+
- Compliance school: government, universities, and granting agencies have embraced Open Science and are mandating some elements (e.g., data sharing with publications)
+
+
+
+
+
+
+
+
Discussion Questions¶
+Which of the pillars of Open Science is nearest to your own heart?
+Open Access Publications
+Open Data
+Open Educational Resources
+Open Methodology
+Open Peer Review
+Open Source Software
+Are any of the pillars more important than the others?
+Are there any pillars not identified that you think should be considered?
+What characteristics might a paper, project, lab group require to qualify as doing Open Science
+What are some barriers to you, your lab group, or your domain doing Open Science?
+What motivates you to do Open Science?
+Do you feel that you fall into a particular "school"? If so, which one, and why?
+Are there any motivating factors for doing Open Science that don't fit into this framework?
+
+
+
+
+
Recommended Open Science Communities¶
+ +
+  +
+  +
+  +
+ Open Scholarship Grassroots Community Networks
+International Open Science Networks
+-
+
- Center for Scientific Collaboration and Community Engagement (CSCCE) +
- Center for Open Science (COS) +
- Eclipse Science Working Group +
- eLife +
- NumFocus +
- Open Access Working Group +
- Open Research Funders Group +
- Open Science Foundation +
- Open Science Network +
- pyOpenSci +
- R OpenSci +
- Research Data Alliance (RDA) +
- The Turing Way +
- UNESCO Global Open Science Partnership +
- World Wide Web Consortium (W3C) +
US-based Open Science Networks
+-
+
- CI Compass - provides expertise and active support to cyberinfrastructure practitioners at USA NSF Major Facilities in order to accelerate the data lifecycle and ensure the integrity and effectiveness of the cyberinfrastructure upon which research and discovery depend. +
- Earth Science Information Partners (ESIP) Federation - is a 501©(3) nonprofit supported by NASA, NOAA, USGS and 130+ member organizations. +
- Internet2 - is a community providing cloud solutions, research support, and services tailored for Research and Education. +
- Minority Serving Cyberinfrastructure Consortium (MS-CC) envisions a transformational partnership to promote advanced cyberinfrastructure (CI) capabilities on the campuses of Historically Black Colleges and Universities (HBCUs), Hispanic-Serving Institutions (HSIs), Tribal Colleges and Universities (TCUs), and other Minority Serving Institutions (MSIs). +
- NASA Transform to Open Science (TOPS) - coordinates efforts designed to rapidly transform agencies, organizations, and communities for Earth Science +
- OpenScapes - is an approach for doing better science for future us +
- The Quilt - non-profit regional research and education networks collaborate to develop, deploy and operate advanced cyberinfrastructure that enables innovation in research and education. +
Oceania Open Science Networks
+-
+
- New Zealand Open Research Network - New Zealand Open Research Network (NZORN) is a collection of researchers and research-associated workers in New Zealand. +
- Australia & New Zealand Open Research Network - ANZORN is a network of local networks distributed without Australia and New Zealand. +
+
+
+
+
+
Self Assessment¶
+True or False: All research papers published in the top journals, like Science and Nature, are always Open Access?
+Answer
+False
+Major Research journals like Science and Nature have an "Open Access" option when a manuscript is accepted, but they charge an extra fee to the authors to make those papers Open Access.
+These high page costs are exclusionary to the majority of global scientists who cannot afford to front these costs out of pocket.
+This will soon change, at least in the United States. The Executive Branch of the federal government recently mandated that future federally funded research be made Open Access after 2026.
+True or False: an article states all of the research data used in the experiments "are available upon request from the corresponding author(s)," meaning the data are "Open"
+Answer
+False
+In order for research to be open, the data need to be freely available from a digital repository, like Data Dryad, Zenodo.org, or CyVerse.
+Data that are 'available upon request' do not meet the FAIR data principles.
+Using a version control system to host the analysis code and computational notebooks, and including these in your Methods section or Supplementary Materials, is an example of an Open Methodology?
+Answer
+Yes!
+Using a VCS like GitHub or GitLab is a great step towards making your research more reproducible.
+Ways to improve your open methology can include documentation of your physical bench work, and even video recordings and step-by-step guides for every part of your project.
+You are asked to review a paper for an important journal in your field. The editor asks if you're willing to release your identity to the authors, thereby "signing" your review. Is this an example of "Open Peer Review"?
+Answer
+Maybe
+There are many opinions on what 'open-review' should consist of. A reviewer signing their review and releasing their identity to the authors is a step toward a more open process. However, it is far less open than publishing the peer-review reports online next to the final published paper.
+You read a paper where the author(s) wrote their own code and licensed as "Open Source" software for a specific set of scientific tasks which you want to replicate. When you visit their personal website, you find the GitHub repository does not exist (because its now private). You contact the authors asking for access, but they refuse to share it 'due to competing researchers who are seeking to steal their intellectual property". Is the software open source?
+Answer
+No
+Just because an author states they have given their software a permissive software license, does not make the software open source.
+Always make certain there is a LICENSE associated with any software you find on the internet.
+In order for the software to be open, it must follow the Open Source Initiative definition
+

 +
+  +
+  +
+  +
+ 
 , meaning building), green check (, meaning success), or red cross (
, meaning building), green check (, meaning success), or red cross ( , meaning failure).
, meaning failure). +
+  +
+  +
+






















 +
+  +
+









 +
+  +
+ +
+  +
+  +
+* An in-memory, columnar database management system optimized for analytical queries."
+* Open-source and designed for simplicity, speed, and efficiency in processing analytical workloads."
+* Supports SQL standards for easy integration with existing tools and workflows.
+___
+## Install DuckDB
+### With Python set up, you can now install DuckDB using pip. In your terminal or command line, execute:
+```console
+pip install duckdb
+```
+___
+
+
+
+___
+
+### To ensure DuckDB is installed correctly, launch your jupyter Notebook and try importing the DuckDB module:
+
+```python
+pip install duckdb
+```
+### If no errors occur, DuckDB is successfully installed and ready to use.
+___
+### In your Jupyter Notebook, import duckdb by running
+```python
+import duckdb
+```
+### For this tutorial, we will be using a pre-configured Duckdb Database.
+### Let's download the database file:
+```python
+!wget --content-disposition https://arizona.box.com/shared/static/uozg0z86rtdjupwpc7i971xwzuzhp42o.duckdb
+```
+
+### Connect to the database using:
+```python
+conn = duckdb.connect(database='/content/my_database.duckdb', read_only=True)
+```
+### Viewing which tables are available inside the database
+
+```python
+conn.sql("SHOW TABLES;")
+```
+
+```python
+┌───────────────────┐
+│ name │
+│ varchar │
+├───────────────────┤
+│ allergies │
+│ careplans │
+│ conditions │
+│ devices │
+│ encounters │
+│ imaging_studies │
+│ immunizations │
+│ medications │
+│ observations │
+│ organizations │
+│ patients │
+│ payer_transitions │
+│ payers │
+│ procedures │
+│ providers │
+│ supplies │
+├───────────────────┤
+│ 16 rows │
+└───────────────────┘
+```
+### Before running any query, we need to know the columns inside particular tables
+
+```python
+conn.sql("DESCRIBE patients;")
+```
+```python
+┌─────────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├─────────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ Id │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ BIRTHDATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ DEATHDATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ SSN │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ DRIVERS │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PASSPORT │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PREFIX │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ FIRST │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ LAST │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ SUFFIX │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ · │ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │ · │
+│ BIRTHPLACE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ADDRESS │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ CITY │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ STATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ COUNTY │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ZIP │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ LAT │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ LON │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ HEALTHCARE_EXPENSES │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ HEALTHCARE_COVERAGE │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+├─────────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┤
+│ 25 rows (20 shown) 6 columns │
+└─────────────────────────────────────────────────────────────────────────┘
+```
+```python
+conn.sql("DESCRIBE medications;")
+```
+```python
+
+conn.sql("DESCRIBE medications;")
+┌───────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├───────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ START │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ STOP │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PATIENT │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PAYER │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ENCOUNTER │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ CODE │ BIGINT │ YES │ NULL │ NULL │ NULL │
+│ DESCRIPTION │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ BASE_COST │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ PAYER_COVERAGE │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ DISPENSES │ BIGINT │ YES │ NULL │ NULL │ NULL │
+│ TOTALCOST │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ REASONCODE │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ REASONDESCRIPTION │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+├───────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┤
+│ 13 rows 6 columns │
+└───────────────────────────────────────────────────────────────────────┘
+```
+```python
+conn.sql("DESCRIBE immunizations;")
+```
+```python
+┌─────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├─────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ DATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PATIENT │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ENCOUNTER │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ CODE │ BIGINT │ YES │ NULL │ NULL │ NULL │
+│ DESCRIPTION │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ BASE_COST │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+└─────────────┴─────────────┴─────────┴─────────┴─────────┴───────┘
+```
+___
+___
+### Querying Data
+#### **SELECT** - The SELECT statement selects data from a database.
+
+E.g.: Query the patients table and display patient first name, last name, gender, and the city they live in.
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients;")
+```
+```python
+┌─────────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├─────────────────┼────────────────┼─────────┼─────────────┤
+│ Jacinto644 │ Kris249 │ M │ Springfield │
+│ Alva958 │ Krajcik437 │ F │ Walpole │
+│ Jimmie93 │ Harris789 │ F │ Pembroke │
+│ Gregorio366 │ Auer97 │ M │ Boston │
+│ Karyn217 │ Mueller846 │ F │ Colrain │
+│ Jayson808 │ Fadel536 │ M │ Chicopee │
+│ José Eduardo181 │ Gómez206 │ M │ Chicopee │
+│ Milo271 │ Feil794 │ M │ Somerville │
+│ Karyn217 │ Metz686 │ F │ Medfield │
+│ Jeffrey461 │ Greenfelder433 │ M │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Raymond398 │ Kuvalis369 │ M │ Hingham │
+│ Gearldine455 │ Boyer713 │ F │ Boston │
+│ Nichol11 │ Gleichner915 │ F │ Lynn │
+│ Louvenia131 │ Marks830 │ F │ Springfield │
+│ Raymon366 │ Beer512 │ M │ Springfield │
+│ Camelia346 │ Stamm704 │ F │ Dedham │
+│ William805 │ Pacocha935 │ M │ Brockton │
+│ Guillermo498 │ Téllez750 │ M │ Somerville │
+│ Milton509 │ Bailey598 │ M │ Hull │
+│ Cecilia788 │ Wisozk929 │ F │ Worcester │
+├─────────────────┴────────────────┴─────────┴─────────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────┘
+```
+### Count the number of rows in patients table
+### This query will count the total number of rows in the patients table, effectively giving you the total number of patients.
+
+```python
+conn.sql("SELECT COUNT(*) FROM patients;")
+```
+```python
+┌──────────────┐
+│ count_star() │
+│ int64 │
+├──────────────┤
+│ 124150 │
+└──────────────┘
+```
+### **COUNT** and **DISTINCT**
+If you want to count distinct values of a specific column, for example, distinct cities, you can modify the query as follows:
+```python
+conn.sql("SELECT COUNT(DISTINCT CITY) FROM patients;")
+```
+```python
+┌──────────────────────┐
+│ count(DISTINCT CITY) │
+│ int64 │
+├──────────────────────┤
+│ 351 │
+└──────────────────────┘
+```
+### Filtering data based on a condition
+The WHERE clause filters records that fulfill a specified condition.
+e.g: Patients in City = 'Springfield';
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY, PASSPORT FROM patients WHERE city = 'Springfield';")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┬────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │ PASSPORT │
+│ varchar │ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┼────────────┤
+│ Jacinto644 │ Kris249 │ M │ Springfield │ NULL │
+│ Jeffrey461 │ Greenfelder433 │ M │ Springfield │ NULL │
+│ Sabina296 │ Flatley871 │ F │ Springfield │ X85058581X │
+│ Theodora872 │ Johnson679 │ F │ Springfield │ X21164602X │
+│ Lavera253 │ Anderson154 │ F │ Springfield │ X83686992X │
+│ Golden321 │ Pollich983 │ F │ Springfield │ NULL │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │ X58134116X │
+│ Fausto876 │ Bechtelar572 │ M │ Springfield │ NULL │
+│ Talisha682 │ Brakus656 │ F │ Springfield │ X53645004X │
+│ Golden321 │ Durgan499 │ F │ Springfield │ X49016634X │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Carla633 │ Terán294 │ F │ Springfield │ X50212869X │
+│ Ángela136 │ Muñiz642 │ F │ Springfield │ NULL │
+│ Marceline716 │ Kuhlman484 │ F │ Springfield │ X70432308X │
+│ Ana María762 │ Cotto891 │ F │ Springfield │ X4604857X │
+│ Josefina523 │ Vanegas191 │ F │ Springfield │ X85396222X │
+│ Ester635 │ Sevilla788 │ F │ Springfield │ X50601786X │
+│ Tomás404 │ Galarza986 │ M │ Springfield │ X76790663X │
+│ Arturo47 │ Delafuente833 │ M │ Springfield │ X45087804X │
+│ Gilberto712 │ Martínez540 │ M │ Springfield │ X89729075X │
+│ Juanita470 │ Connelly992 │ F │ Springfield │ X71156217X │
+├──────────────┴────────────────┴─────────┴─────────────┴────────────┤
+│ 2814 rows (20 shown) 5 columns │
+└────────────────────────────────────────────────────────────────────┘
+```
+### Multiple conditions in Selection
+e.g 1: Female Patients in City = 'Springfield';
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE city = 'Springfield' AND gender = 'F';")
+```
+```python
+
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE city = 'Springfield' AND gender = 'F';")
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │
+│ Theodora872 │ Johnson679 │ F │ Springfield │
+│ Lavera253 │ Anderson154 │ F │ Springfield │
+│ Golden321 │ Pollich983 │ F │ Springfield │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Golden321 │ Durgan499 │ F │ Springfield │
+│ Jerrie417 │ Gislason620 │ F │ Springfield │
+│ Venus149 │ Hodkiewicz467 │ F │ Springfield │
+│ Refugia211 │ Wintheiser220 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Tisha655 │ Renner328 │ F │ Springfield │
+│ Mona85 │ Senger904 │ F │ Springfield │
+│ Isabel214 │ Camacho176 │ F │ Springfield │
+│ Carla633 │ Terán294 │ F │ Springfield │
+│ Ángela136 │ Muñiz642 │ F │ Springfield │
+│ Marceline716 │ Kuhlman484 │ F │ Springfield │
+│ Ana María762 │ Cotto891 │ F │ Springfield │
+│ Josefina523 │ Vanegas191 │ F │ Springfield │
+│ Ester635 │ Sevilla788 │ F │ Springfield │
+│ Juanita470 │ Connelly992 │ F │ Springfield │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 1351 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+### Nested selection
+e.g. Female patients from City Springfield or Boston
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE gender = 'F' AND (city = 'Springfield' OR city = 'Boston');")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │
+│ Theodora872 │ Johnson679 │ F │ Springfield │
+│ Lavera253 │ Anderson154 │ F │ Springfield │
+│ Golden321 │ Pollich983 │ F │ Springfield │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Golden321 │ Durgan499 │ F │ Springfield │
+│ Jerrie417 │ Gislason620 │ F │ Springfield │
+│ Venus149 │ Hodkiewicz467 │ F │ Springfield │
+│ Refugia211 │ Wintheiser220 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Freida957 │ Hand679 │ F │ Boston │
+│ Clora637 │ Rempel203 │ F │ Boston │
+│ Bonny428 │ Turner526 │ F │ Boston │
+│ Tien590 │ Gaylord332 │ F │ Boston │
+│ Arlyne429 │ Deckow585 │ F │ Boston │
+│ Doreen575 │ Johnson679 │ F │ Boston │
+│ Kym935 │ Hayes766 │ F │ Boston │
+│ Julia241 │ Nevárez403 │ F │ Boston │
+│ Ja391 │ Murray856 │ F │ Boston │
+│ Creola518 │ Spinka232 │ F │ Boston │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 7191 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+#### Alternative
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE gender = 'F' AND (city IN ('Springfield','Boston'));")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │
+│ Theodora872 │ Johnson679 │ F │ Springfield │
+│ Lavera253 │ Anderson154 │ F │ Springfield │
+│ Golden321 │ Pollich983 │ F │ Springfield │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Golden321 │ Durgan499 │ F │ Springfield │
+│ Jerrie417 │ Gislason620 │ F │ Springfield │
+│ Venus149 │ Hodkiewicz467 │ F │ Springfield │
+│ Refugia211 │ Wintheiser220 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Freida957 │ Hand679 │ F │ Boston │
+│ Clora637 │ Rempel203 │ F │ Boston │
+│ Bonny428 │ Turner526 │ F │ Boston │
+│ Tien590 │ Gaylord332 │ F │ Boston │
+│ Arlyne429 │ Deckow585 │ F │ Boston │
+│ Doreen575 │ Johnson679 │ F │ Boston │
+│ Kym935 │ Hayes766 │ F │ Boston │
+│ Julia241 │ Nevárez403 │ F │ Boston │
+│ Ja391 │ Murray856 │ F │ Boston │
+│ Creola518 │ Spinka232 │ F │ Boston │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 7191 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+### Filter on missing data
+* Filtering on missing data is crucial for maintaining data integrity and ensuring accurate analysis by identifying and handling incomplete records effectively.
+* Retrieve the first name, last name, gender, and city for all patients who are from Springfield and have a passport number recorded (i.e., the passport field is not empty).
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY, PASSPORT FROM patients WHERE city = 'Springfield' AND PASSPORT IS NOT NULL;")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┬────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │ PASSPORT │
+│ varchar │ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┼────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │ X85058581X │
+│ Theodora872 │ Johnson679 │ F │ Springfield │ X21164602X │
+│ Lavera253 │ Anderson154 │ F │ Springfield │ X83686992X │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │ X58134116X │
+│ Talisha682 │ Brakus656 │ F │ Springfield │ X53645004X │
+│ Golden321 │ Durgan499 │ F │ Springfield │ X49016634X │
+│ Ty725 │ Schmeler639 │ M │ Springfield │ X22960735X │
+│ Pilar644 │ Pouros728 │ F │ Springfield │ X21434326X │
+│ Georgette866 │ Stark857 │ F │ Springfield │ X84034866X │
+│ Annika454 │ Gutmann970 │ F │ Springfield │ X3275916X │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Isabel214 │ Camacho176 │ F │ Springfield │ X52173808X │
+│ Carla633 │ Terán294 │ F │ Springfield │ X50212869X │
+│ Marceline716 │ Kuhlman484 │ F │ Springfield │ X70432308X │
+│ Ana María762 │ Cotto891 │ F │ Springfield │ X4604857X │
+│ Josefina523 │ Vanegas191 │ F │ Springfield │ X85396222X │
+│ Ester635 │ Sevilla788 │ F │ Springfield │ X50601786X │
+│ Tomás404 │ Galarza986 │ M │ Springfield │ X76790663X │
+│ Arturo47 │ Delafuente833 │ M │ Springfield │ X45087804X │
+│ Gilberto712 │ Martínez540 │ M │ Springfield │ X89729075X │
+│ Juanita470 │ Connelly992 │ F │ Springfield │ X71156217X │
+├──────────────┴────────────────┴─────────┴─────────────┴────────────┤
+│ 2221 rows (20 shown) 5 columns │
+└────────────────────────────────────────────────────────────────────┘
+```
+### Filter and select in numeric range
+Patients in City = 'Springfield' where the HEALTHCARE_EXPENSES between 1.5M and 2M
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE city = 'Springfield' AND HEALTHCARE_EXPENSES BETWEEN 1500000 AND 2000000;")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Jerold208 │ Harber290 │ M │ Springfield │
+│ Dean966 │ Tillman293 │ M │ Springfield │
+│ Orval846 │ Cartwright189 │ M │ Springfield │
+│ Jacinto644 │ Abernathy524 │ M │ Springfield │
+│ Bethel526 │ Satterfield305 │ F │ Springfield │
+│ Dorene845 │ Botsford977 │ F │ Springfield │
+│ Lula998 │ Langosh790 │ F │ Springfield │
+│ Deeanna316 │ Koss676 │ F │ Springfield │
+│ Loriann967 │ Torp761 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Ashley34 │ Murazik203 │ M │ Springfield │
+│ Ulysses632 │ Donnelly343 │ M │ Springfield │
+│ Horace32 │ Hammes673 │ M │ Springfield │
+│ Roman389 │ Lubowitz58 │ M │ Springfield │
+│ Jerald662 │ Grady603 │ M │ Springfield │
+│ Damien170 │ Hoppe518 │ M │ Springfield │
+│ Charley358 │ Vandervort697 │ M │ Springfield │
+│ Carla633 │ Terán294 │ F │ Springfield │
+│ Ana María762 │ Cotto891 │ F │ Springfield │
+│ Juanita470 │ Connelly992 │ F │ Springfield │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 241 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+### LIMIT
+- LIMIT specifies the maximum number of records the query will return.
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients LIMIT 20;")
+```
+```python
+
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients LIMIT 20;")
+┌─────────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├─────────────────┼────────────────┼─────────┼─────────────┤
+│ Jacinto644 │ Kris249 │ M │ Springfield │
+│ Alva958 │ Krajcik437 │ F │ Walpole │
+│ Jimmie93 │ Harris789 │ F │ Pembroke │
+│ Gregorio366 │ Auer97 │ M │ Boston │
+│ Karyn217 │ Mueller846 │ F │ Colrain │
+│ Jayson808 │ Fadel536 │ M │ Chicopee │
+│ José Eduardo181 │ Gómez206 │ M │ Chicopee │
+│ Milo271 │ Feil794 │ M │ Somerville │
+│ Karyn217 │ Metz686 │ F │ Medfield │
+│ Jeffrey461 │ Greenfelder433 │ M │ Springfield │
+│ Mariana775 │ Gulgowski816 │ F │ Lowell │
+│ Leann224 │ Deckow585 │ F │ Needham │
+│ Isabel214 │ Hinojosa147 │ F │ Fall River │
+│ Christal240 │ Brown30 │ F │ Boston │
+│ Carmelia328 │ Konopelski743 │ F │ Ashland │
+│ Raye931 │ Wyman904 │ F │ Quincy │
+│ Lisbeth69 │ Rowe323 │ F │ Malden │
+│ Amada498 │ Spinka232 │ F │ Foxborough │
+│ Cythia210 │ Reichel38 │ F │ Peabody │
+│ María Soledad68 │ Aparicio848 │ F │ Boston │
+├─────────────────┴────────────────┴─────────┴─────────────┤
+│ 20 rows 4 columns │
+└──────────────────────────────────────────────────────────┘
+```
+### ORDER BY and ASC
+This query returns the first 20 patients from the patients table, ordered alphabetically by their last name.
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients ORDER BY LAST ASC LIMIT 20;")
+```
+```python
+┌────────────────┬───────────┬─────────┬──────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├────────────────┼───────────┼─────────┼──────────────┤
+│ Jc393 │ Abbott774 │ M │ Westfield │
+│ Lloyd546 │ Abbott774 │ M │ Northampton │
+│ Charles364 │ Abbott774 │ M │ Brockton │
+│ Lelia627 │ Abbott774 │ F │ Gloucester │
+│ Devorah937 │ Abbott774 │ F │ Cambridge │
+│ Warren653 │ Abbott774 │ M │ Tyngsborough │
+│ Rhona164 │ Abbott774 │ F │ Stoneham │
+│ Jackqueline794 │ Abbott774 │ F │ Randolph │
+│ Willa615 │ Abbott774 │ F │ Dartmouth │
+│ Cyril535 │ Abbott774 │ M │ Millis │
+│ Lorette239 │ Abbott774 │ F │ Dennis │
+│ Jimmy858 │ Abbott774 │ M │ Lowell │
+│ Laine739 │ Abbott774 │ F │ Agawam │
+│ Bernetta267 │ Abbott774 │ F │ Ware │
+│ Lauri399 │ Abbott774 │ F │ Springfield │
+│ Miesha237 │ Abbott774 │ F │ Stoneham │
+│ Darrin898 │ Abbott774 │ M │ Newton │
+│ Grant908 │ Abbott774 │ M │ Arlington │
+│ Arden380 │ Abbott774 │ M │ Worcester │
+│ German382 │ Abbott774 │ M │ Taunton │
+├────────────────┴───────────┴─────────┴──────────────┤
+│ 20 rows 4 columns │
+└─────────────────────────────────────────────────────┘
+```
+### ORDER BY and DESC
+This query returns the first 20 patients from the patients table, ordered reverse alphabetically by their first name.
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients ORDER BY FIRST DESC LIMIT 20;")
+```
+```python
+┌──────────┬────────────────┬─────────┬────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────┼────────────────┼─────────┼────────────┤
+│ Óscar156 │ Ballesteros368 │ M │ Taunton │
+│ Óscar156 │ Curiel392 │ M │ Swampscott │
+│ Óscar156 │ Zelaya592 │ M │ Lynn │
+│ Óscar156 │ Puente961 │ M │ Greenfield │
+│ Óscar156 │ Olivas524 │ M │ Norwell │
+│ Óscar156 │ Rivero165 │ M │ Lancaster │
+│ Óscar156 │ Canales95 │ M │ Hamilton │
+│ Óscar156 │ Romero158 │ M │ Boston │
+│ Óscar156 │ Garza151 │ M │ Longmeadow │
+│ Óscar156 │ Delgado712 │ M │ Haverhill │
+│ Óscar156 │ Ureña88 │ M │ Bedford │
+│ Óscar156 │ Henríquez109 │ M │ Boxborough │
+│ Óscar156 │ Guerrero997 │ M │ Boston │
+│ Óscar156 │ Ojeda263 │ M │ Boston │
+│ Óscar156 │ Meléndez48 │ M │ Newton │
+│ Óscar156 │ Muro989 │ M │ Boston │
+│ Óscar156 │ Rendón540 │ M │ Wilbraham │
+│ Óscar156 │ Santacruz647 │ M │ Winthrop │
+│ Óscar156 │ Santacruz647 │ M │ Boston │
+│ Óscar156 │ Otero621 │ M │ Boston │
+├──────────┴────────────────┴─────────┴────────────┤
+│ 20 rows 4 columns │
+└──────────────────────────────────────────────────┘
+```
+## Aggregating Data using GROUP BY
+#### Counting Patients by City
+
+```python
+conn.sql("SELECT CITY, COUNT(*) AS patient_count FROM patients GROUP BY CITY;") ## each run gives random order
+```
+```python
+┌─────────────┬───────────────┐
+│ CITY │ patient_count │
+│ varchar │ int64 │
+├─────────────┼───────────────┤
+│ Dartmouth │ 656 │
+│ Fitchburg │ 743 │
+│ Plymouth │ 972 │
+│ Worcester │ 3263 │
+│ Beverly │ 778 │
+│ Westfield │ 781 │
+│ Hingham │ 431 │
+│ Rochester │ 103 │
+│ Sandwich │ 387 │
+│ Watertown │ 534 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Ashfield │ 29 │
+│ Pelham │ 26 │
+│ Hancock │ 22 │
+│ Monterey │ 13 │
+│ Richmond │ 23 │
+│ Hinsdale │ 30 │
+│ Middlefield │ 11 │
+│ Westhampton │ 14 │
+│ Sandisfield │ 12 │
+│ Monroe │ 2 │
+├─────────────┴───────────────┤
+│ 351 rows (20 shown) │
+└─────────────────────────────┘
+```
+### Counting Patients by City and sorting them highest to lowest
+```python
+conn.sql("SELECT CITY, COUNT(*) AS patient_count FROM patients GROUP BY CITY ORDER BY patient_count DESC;")
+```
+```python
+┌─────────────┬───────────────┐
+│ CITY │ patient_count │
+│ varchar │ int64 │
+├─────────────┼───────────────┤
+│ Boston │ 11496 │
+│ Worcester │ 3263 │
+│ Springfield │ 2814 │
+│ Cambridge │ 2044 │
+│ Lowell │ 2027 │
+│ Brockton │ 1833 │
+│ Lynn │ 1714 │
+│ New Bedford │ 1706 │
+│ Quincy │ 1698 │
+│ Newton │ 1695 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Blandford │ 10 │
+│ Heath │ 8 │
+│ Tolland │ 7 │
+│ Hawley │ 6 │
+│ Alford │ 5 │
+│ New Ashford │ 4 │
+│ Aquinnah │ 4 │
+│ Tyringham │ 3 │
+│ Monroe │ 2 │
+│ Gosnold │ 1 │
+├─────────────┴───────────────┤
+│ 351 rows (20 shown) │
+└─────────────────────────────┘
+```
+### Total Number of Patients by Gender in Each City
+```python
+conn.sql("SELECT CITY, GENDER, COUNT(*) AS total_patients FROM patients GROUP BY CITY, GENDER ORDER BY total_patients DESC LIMIT 10;")
+```
+```python
+┌─────────────┬─────────┬────────────────┐
+│ CITY │ GENDER │ total_patients │
+│ varchar │ varchar │ int64 │
+├─────────────┼─────────┼────────────────┤
+│ Boston │ F │ 5840 │
+│ Boston │ M │ 5656 │
+│ Worcester │ M │ 1694 │
+│ Worcester │ F │ 1569 │
+│ Springfield │ M │ 1463 │
+│ Springfield │ F │ 1351 │
+│ Cambridge │ M │ 1023 │
+│ Cambridge │ F │ 1021 │
+│ Lowell │ F │ 1015 │
+│ Lowell │ M │ 1012 │
+├─────────────┴─────────┴────────────────┤
+│ 10 rows 3 columns │
+└────────────────────────────────────────┘
+```
+Lets look at other Tables - observations and immunizations
+```python
+conn.sql("SELECT PATIENT,ENCOUNTER,CODE, DESCRIPTION, VALUE from observations;")
+```
+```python
+┌──────────────────────┬──────────────────────┬─────────┬───────────────────────────────────────────────┬──────────────┐
+│ PATIENT │ ENCOUNTER │ CODE │ DESCRIPTION │ VALUE │
+│ varchar │ varchar │ varchar │ varchar │ varchar │
+├──────────────────────┼──────────────────────┼─────────┼───────────────────────────────────────────────┼──────────────┤
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8302-2 │ Body Height │ 82.7 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 72514-3 │ Pain severity - 0-10 verbal numeric rating … │ 2.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 29463-7 │ Body Weight │ 11.5 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 77606-2 │ Weight-for-length Per age and sex │ 47.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 9843-4 │ Head Occipital-frontal circumference │ 46.9 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8462-4 │ Diastolic Blood Pressure │ 76.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8480-6 │ Systolic Blood Pressure │ 107.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8867-4 │ Heart rate │ 68.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 9279-1 │ Respiratory rate │ 13.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 72166-2 │ Tobacco smoking status NHIS │ Never smoker │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2160-0 │ Creatinine [Mass/volume] in Serum or Plasma │ 2.9 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 17861-6 │ Calcium [Mass/volume] in Serum or Plasma │ 9.2 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2951-2 │ Sodium [Moles/volume] in Serum or Plasma │ 143.8 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2823-3 │ Potassium [Moles/volume] in Serum or Plasma │ 4.3 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2075-0 │ Chloride [Moles/volume] in Serum or Plasma │ 106.4 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2028-9 │ Carbon dioxide total [Moles/volume] in Ser… │ 22.9 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 33914-3 │ Glomerular filtration rate/1.73 sq M.predic… │ 9.9 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2885-2 │ Protein [Mass/volume] in Serum or Plasma │ 5.7 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 1751-7 │ Albumin [Mass/volume] in Serum or Plasma │ 5.2 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 1975-2 │ Bilirubin.total [Mass/volume] in Serum or P… │ 14.2 │
+├──────────────────────┴──────────────────────┴─────────┴───────────────────────────────────────────────┴──────────────┤
+│ ? rows (>9999 rows, 20 shown) 5 columns │
+└──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+#### Average Heart Rate by Patient
+
+```python
+conn.sql("SELECT PATIENT, ROUND(AVG(CAST(VALUE AS DECIMAL(10,6))), 2) AS average_heart_rate FROM observations WHERE DESCRIPTION LIKE '%Heart rate%' GROUP BY PATIENT ORDER BY average_heart_rate DESC LIMIT 20;")
+```
+```python
+┌──────────────────────────────────────┬────────────────────┐
+│ PATIENT │ average_heart_rate │
+│ varchar │ double │
+├──────────────────────────────────────┼────────────────────┤
+│ ff2209f2-a4c9-4737-9aa7-adc8d71b6961 │ 200.0 │
+│ 8cea2d7e-6227-4924-a340-18eafb8564ac │ 200.0 │
+│ 6ec823e7-839e-4491-8b6f-6b275951b456 │ 200.0 │
+│ 0191f8aa-96dc-41e6-b718-312686a7a867 │ 200.0 │
+│ c10db4e1-de51-495f-98ab-8322c6d9550a │ 200.0 │
+│ 46ea7226-63f6-404f-bbed-904ea39f706d │ 199.9 │
+│ ec1ce32c-57a3-44c4-8f64-e050f2f94e03 │ 199.9 │
+│ a05e3bd1-1c51-453a-9969-646cb0168d23 │ 199.9 │
+│ 9cc52681-4446-4e4a-80be-63418dd06e66 │ 199.9 │
+│ b0112890-a24a-4564-b2b3-9b77879e5e79 │ 199.9 │
+│ 18f53179-2449-47af-b4da-a9e51095076f │ 199.9 │
+│ c3d165d2-597e-45a8-8331-f3338ce19fdf │ 199.9 │
+│ 04dd3439-2116-4f9a-b40e-a84395cf6f21 │ 199.9 │
+│ 371e14a1-31bd-4407-bdbb-4078781fde05 │ 199.8 │
+│ 3e5c63bf-65fe-48bb-8361-6b21a909888a │ 199.8 │
+│ 423f79cf-f7b8-41cd-bb3c-8459084f88eb │ 199.8 │
+│ 2d4f57ee-4001-44f8-9a94-c1885f26b408 │ 199.8 │
+│ bd6784c0-3529-4a12-87bd-746affb17739 │ 199.8 │
+│ ef43848b-a4f1-4c45-a106-5928e29b72f2 │ 199.7 │
+│ 1d050245-097c-44a5-b29b-6a18a22731e9 │ 199.7 │
+├──────────────────────────────────────┴────────────────────┤
+│ 20 rows 2 columns │
+└───────────────────────────────────────────────────────────┘
+```
+#### Count of Heart Rate Records by Patient
+```python
+conn.sql("SELECT PATIENT, COUNT(*) AS heart_rate_records FROM observations WHERE DESCRIPTION LIKE '%Heart rate%' GROUP BY PATIENT ORDER BY heart_rate_records DESC LIMIT 20;")
+```
+```python
+┌──────────────────────────────────────┬────────────────────┐
+│ PATIENT │ heart_rate_records │
+│ varchar │ int64 │
+├──────────────────────────────────────┼────────────────────┤
+│ 3e21a156-da54-4fb7-815e-550fdf4afbbd │ 33 │
+│ ffbf0392-1643-4b05-819f-489072c8c4d4 │ 30 │
+│ b9ef6005-438e-4b47-afaf-9dba32184adf │ 30 │
+│ f8080701-2b5c-4128-9a7f-9a098b737b27 │ 30 │
+│ a2172279-3d63-4d14-867d-1a39d9280690 │ 30 │
+│ 01a5a5a6-ef7b-42ba-899c-de66e1b1e27e │ 30 │
+│ 62572c44-a802-40d1-8d60-d02bc287d548 │ 30 │
+│ 445953fd-15fa-424e-9926-f93ebf1bca7a │ 30 │
+│ c3ea3c46-f8d1-4abe-8fb2-8fc8f86b4ef4 │ 29 │
+│ bd18ba0d-2e65-4427-98f1-cbfc09dbaa33 │ 29 │
+│ 5736b489-0e15-4693-b9d6-05192a702a2d │ 29 │
+│ b5a0b060-96ae-44d5-bd10-f532a445009b │ 29 │
+│ ee9b44a1-36c6-456e-b77c-cd200611ca0f │ 29 │
+│ 0d5290dd-a3e3-4a81-91d5-6e722e58ec3e │ 29 │
+│ 54ec8b8e-fe2a-4cf5-a087-1a65df1ab1b8 │ 29 │
+│ e658626d-0f72-4570-8071-2dd63827a24c │ 29 │
+│ ec3f3da2-09a1-43c7-8cae-ed34847d2534 │ 28 │
+│ c7bd54b1-e036-4702-9062-acfa3677a6e8 │ 28 │
+│ 8b6ee161-eb5f-413a-8e86-f5b3adb3f5ff │ 28 │
+│ 03c1210e-d145-4aed-9522-14cff41b3c28 │ 28 │
+├──────────────────────────────────────┴────────────────────┤
+│ 20 rows 2 columns │
+└───────────────────────────────────────────────────────────┘
+```
+#### Total Cost of Vaccinations per Patient
+```python
+conn.sql("SELECT PATIENT, SUM(BASE_COST) AS total_cost FROM immunizations GROUP BY PATIENT ORDER BY total_cost DESC;")
+```
+```python
+┌──────────────────────────────────────┬────────────┐
+│ PATIENT │ total_cost │
+│ varchar │ double │
+├──────────────────────────────────────┼────────────┤
+│ 8eacdc17-de2f-4024-88bd-976401b30979 │ 2810.4 │
+│ bb14de6a-c77d-44bb-a3a8-338a6a246520 │ 2810.4 │
+│ a5b60d43-9776-4811-badf-67d49edbc175 │ 2810.4 │
+│ 1c8a4026-5dbe-478a-b647-ecb605775977 │ 2810.4 │
+│ 3e38e20b-6dd8-44fe-9ed3-81e1e3d8e2ab │ 2810.4 │
+│ e23c7c68-0319-41f5-90d9-1db160321a94 │ 2810.4 │
+│ 49e7d878-93a6-4e71-971b-213abacf2235 │ 2810.4 │
+│ 7ded4116-1783-4a93-9bb2-7a65370f55f5 │ 2810.4 │
+│ 594660e4-6a02-4e3c-a8b9-2c61d4d4af60 │ 2810.4 │
+│ fee1edbd-277d-4b12-901e-60b6135cf877 │ 2810.4 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ 594d1ecb-b1a4-4bad-9ccd-5cebceebd6f5 │ 281.04 │
+│ 456fa9f9-7eb7-4316-a74b-5a9ff87f8fc5 │ 281.04 │
+│ 52568520-7163-429d-bc90-fc245debb697 │ 281.04 │
+│ 30a16a19-ee93-4bfd-b1ed-bba90de36266 │ 281.04 │
+│ 0ef633b0-2996-44c5-936c-600820310223 │ 281.04 │
+│ 613319c8-2b76-494b-86b8-e776a2ee5e0b │ 281.04 │
+│ 4dceeb57-4fe3-4c32-a748-621df5ce3c30 │ 281.04 │
+│ 93d590e8-9f3e-4741-bee2-9b8e9e8b21e7 │ 281.04 │
+│ bad20ffc-48f9-4a7d-a2be-39a50fa4e0ac │ 281.04 │
+│ 3cf75ec3-5e1c-4783-bb20-282a8cf66f30 │ 281.04 │
+├──────────────────────────────────────┴────────────┤
+│ ? rows (>9999 rows, 20 shown) 2 columns │
+└───────────────────────────────────────────────────┘
+```
+#### Top 10 Most Common Vaccine Administered
+```python
+conn.sql("SELECT DESCRIPTION, COUNT(*) AS count FROM immunizations GROUP BY DESCRIPTION ORDER BY count DESC LIMIT 10;")
+```
+```python
+┌────────────────────────────────────────────────────┬────────┐
+│ DESCRIPTION │ count │
+│ varchar │ int64 │
+├────────────────────────────────────────────────────┼────────┤
+│ Influenza seasonal injectable preservative free │ 106564 │
+│ Td (adult) preservative free │ 9815 │
+│ Pneumococcal conjugate PCV 13 │ 5747 │
+│ DTaP │ 5735 │
+│ IPV │ 4962 │
+│ meningococcal MCV4P │ 4010 │
+│ Hib (PRP-OMP) │ 3615 │
+│ HPV quadrivalent │ 3494 │
+│ Hep B adolescent or pediatric │ 3490 │
+│ zoster │ 3469 │
+├────────────────────────────────────────────────────┴────────┤
+│ 10 rows 2 columns │
+└─────────────────────────────────────────────────────────────┘
+```
+## JOINS
+#### SQL joins are used to combine rows from two or more tables, based on a related column between them. There are several types of joins in SQL, each serving different purposes depending on how you want to combine your data.
+
+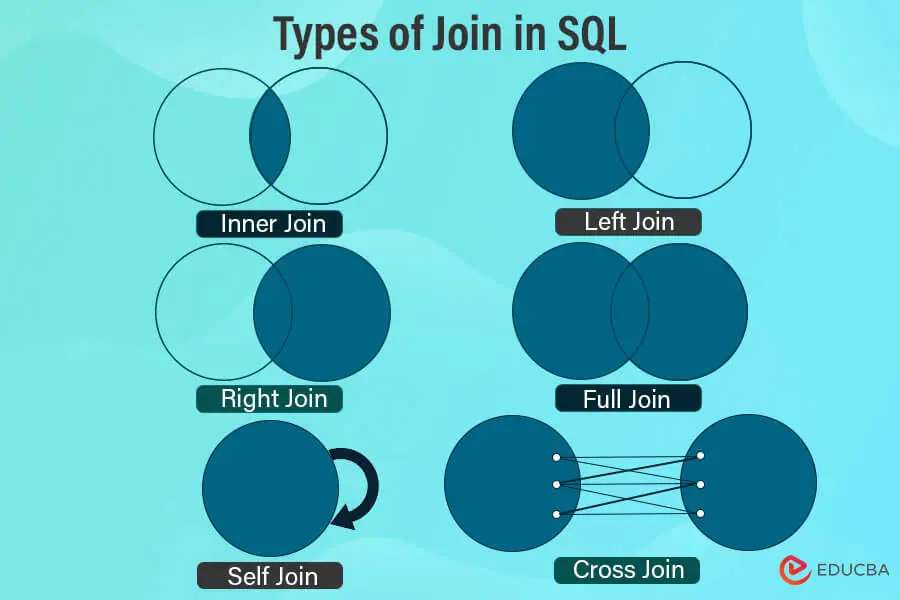
+ - credit: educba.com
+
+___
+
+For the following examples: Left table: Patients and Right Table: Immunizations
+
+__
+
+### INNER JOIN
+- The Inner join returns rows when there is at least one match in both tables. If there is no match, the rows are not returned.
+
+- Find Matching Records: Question: Which medical treatments have been administered to patients, including the patient's name and the cost of each treatment?
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM immunizations INNER JOIN patients ON immunizations.PATIENT = patients.Id ORDER BY patients.FIRST ASC;")
+```
+```python
+┌──────────┬───────────────┬────────────────────────────────────────────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├──────────┼───────────────┼────────────────────────────────────────────────────┼───────────┤
+│ Aaron697 │ Cummings51 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Anderson154 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Aaron697 │ Hartmann983 │ Hep B adolescent or pediatric │ 140.52 │
+│ Aaron697 │ Thompson596 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Rodriguez71 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Deckow585 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Swaniawski813 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Beer512 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Ullrich385 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Ariel183 │ Schuster709 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Mohr916 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Waters156 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Will178 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Leffler128 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Bode78 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Grady603 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Funk324 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ McGlynn426 │ Td (adult) preservative free │ 140.52 │
+│ Ariel183 │ Boehm581 │ Influenza seasonal injectable preservative free │ 140.52 │
+├──────────┴───────────────┴────────────────────────────────────────────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└───────────────────────────────────────────────────────────────────────────────────────────┘
+```
+
+### LEFT JOIN
+- It returns all rows from the left table, and the matched rows from the right table. The result is NULL from the right side, if there is no match.
+Question: For all patients, what treatments have they received, if any?
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients LEFT JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY patients.FIRST ASC;")
+```
+```python
+
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients LEFT JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY patients.FIRST ASC;")
+┌───────────┬─────────────┬────────────────────────────────────────────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├───────────┼─────────────┼────────────────────────────────────────────────────┼───────────┤
+│ Aaron697 │ Legros616 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Deckow585 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Thompson596 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Auer97 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Td (adult) preservative free │ 140.52 │
+│ Aaron697 │ Volkman526 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Hartmann983 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Orn563 │ NULL │ NULL │
+│ Aaron697 │ Hartmann983 │ IPV │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Antonia30 │ Goodwin327 │ HPV quadrivalent │ 140.52 │
+│ Antonia30 │ Funk324 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ West559 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Lowe577 │ NULL │ NULL │
+│ Antonia30 │ D'Amore443 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Montes106 │ meningococcal MCV4P │ 140.52 │
+│ Antonia30 │ González124 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Franecki195 │ zoster │ 140.52 │
+│ Antonia30 │ Rojo930 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Puente961 │ HPV quadrivalent │ 140.52 │
+├───────────┴─────────────┴────────────────────────────────────────────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────────────────────────────────────┘
+```
+### RIGHT JOIN
+- It returns all rows from the right table, and the matched rows from the left table. The result is NULL from the left side, if there is no match.
+Question: For all Immunizations recorded, which patients received them, if identifiable?
+
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients RIGHT JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY immunizations.DESCRIPTION;")
+```
+```python
+┌────────────────┬──────────────┬──────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├────────────────┼──────────────┼──────────────┼───────────┤
+│ Shantay950 │ Quitzon246 │ DTaP │ 140.52 │
+│ Cleveland582 │ VonRueden376 │ DTaP │ 140.52 │
+│ Flo729 │ Quigley282 │ DTaP │ 140.52 │
+│ Flo729 │ Quigley282 │ DTaP │ 140.52 │
+│ Werner409 │ Schaden604 │ DTaP │ 140.52 │
+│ Pasty639 │ Ortiz186 │ DTaP │ 140.52 │
+│ Pasty639 │ Ortiz186 │ DTaP │ 140.52 │
+│ Angelica442 │ Kovacek682 │ DTaP │ 140.52 │
+│ Cathie710 │ Hegmann834 │ DTaP │ 140.52 │
+│ Pasty639 │ Ortiz186 │ DTaP │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Art115 │ Roberts511 │ Hep A adult │ 140.52 │
+│ Christopher407 │ Davis923 │ Hep A adult │ 140.52 │
+│ Michiko564 │ Dooley940 │ Hep A adult │ 140.52 │
+│ Celine582 │ Sipes176 │ Hep A adult │ 140.52 │
+│ Darci883 │ Miller503 │ Hep A adult │ 140.52 │
+│ Ethan766 │ Morar593 │ Hep A adult │ 140.52 │
+│ Georgiann138 │ Heathcote539 │ Hep A adult │ 140.52 │
+│ Herbert830 │ Wolff180 │ Hep A adult │ 140.52 │
+│ Ismael683 │ King743 │ Hep A adult │ 140.52 │
+│ Ellsworth48 │ Mertz280 │ Hep A adult │ 140.52 │
+├────────────────┴──────────────┴──────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────┘
+```
+### FULL OUTER JOIN
+- It returns rows when there is a match in one of the tables. It effectively combines the results of both LEFT JOIN and RIGHT JOIN.
+Question: What is the complete list of patients and their Immunizations, including those without recorded treatments or identifiable patients?
+
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients FULL OUTER JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY patients.FIRST ASC;")
+```
+```python
+┌───────────┬─────────────┬────────────────────────────────────────────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├───────────┼─────────────┼────────────────────────────────────────────────────┼───────────┤
+│ Aaron697 │ Legros616 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Deckow585 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Thompson596 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Auer97 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Td (adult) preservative free │ 140.52 │
+│ Aaron697 │ Volkman526 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Hartmann983 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Orn563 │ NULL │ NULL │
+│ Aaron697 │ Hartmann983 │ IPV │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Antonia30 │ Goodwin327 │ HPV quadrivalent │ 140.52 │
+│ Antonia30 │ Funk324 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ West559 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Lowe577 │ NULL │ NULL │
+│ Antonia30 │ D'Amore443 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Montes106 │ meningococcal MCV4P │ 140.52 │
+│ Antonia30 │ González124 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Franecki195 │ zoster │ 140.52 │
+│ Antonia30 │ Rojo930 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Puente961 │ HPV quadrivalent │ 140.52 │
+├───────────┴─────────────┴────────────────────────────────────────────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────────────────────────────────────┘
+```
+### CROSS JOIN
+- It returns a Cartesian product of the two tables, i.e., it returns rows combining each row from the first table with each row from the second table.
+Question: What are all possible combinations of patients and immunizations?
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION FROM patients CROSS JOIN immunizations;")
+```
+```python
+┌───────────┬─────────────┬────────────────────────────────────────────────────┐
+│ FIRST │ LAST │ DESCRIPTION │
+│ varchar │ varchar │ varchar │
+├───────────┼─────────────┼────────────────────────────────────────────────────┤
+│ Tammy740 │ Ernser583 │ Influenza seasonal injectable preservative free │
+│ Tammy740 │ Ernser583 │ Hep A ped/adol 2 dose │
+│ Tammy740 │ Ernser583 │ Influenza seasonal injectable preservative free │
+│ Tammy740 │ Ernser583 │ Influenza seasonal injectable preservative free │
+│ Tammy740 │ Ernser583 │ meningococcal MCV4P │
+│ Tammy740 │ Ernser583 │ Hep B adolescent or pediatric │
+│ Tammy740 │ Ernser583 │ Hep B adolescent or pediatric │
+│ Tammy740 │ Ernser583 │ Hib (PRP-OMP) │
+│ Tammy740 │ Ernser583 │ rotavirus monovalent │
+│ Tammy740 │ Ernser583 │ IPV │
+│ · │ · │ · │
+│ · │ · │ · │
+│ · │ · │ · │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ zoster │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+├───────────┴─────────────┴────────────────────────────────────────────────────┤
+│ ? rows (>9999 rows, 20 shown) 3 columns │
+└──────────────────────────────────────────────────────────────────────────────┘
+```
+### SELF JOIN
+- It is not a different type of join, but it's a regular join used to join a table to itself. It's useful for queries where you need to compare rows within the same table.
+Question: Which pair of patients are from the same city?
+```python
+conn.sql("SELECT A.FIRST AS FirstPatient, B.FIRST AS SecondPatient, A.CITY FROM patients A, patients B WHERE A.CITY = B.CITY AND A.Id != B.Id ORDER BY A.CITY ASC;")
+```
+```python
+┌──────────────┬───────────────┬──────────┐
+│ FirstPatient │ SecondPatient │ CITY │
+│ varchar │ varchar │ varchar │
+├──────────────┼───────────────┼──────────┤
+│ Susan422 │ Stefani254 │ Abington │
+│ Jennell254 │ Lionel365 │ Abington │
+│ Robbie31 │ Antonio44 │ Abington │
+│ Joe656 │ Antonio44 │ Abington │
+│ Susan422 │ Stevie682 │ Abington │
+│ Shaun461 │ Stevie682 │ Abington │
+│ Shaun461 │ Weston546 │ Abington │
+│ Susan422 │ Elwood28 │ Abington │
+│ Susan422 │ Daria61 │ Abington │
+│ Shaun461 │ Lita714 │ Abington │
+│ · │ · │ · │
+│ · │ · │ · │
+│ · │ · │ · │
+│ Elane105 │ Conrad619 │ Abington │
+│ Logan497 │ Conrad619 │ Abington │
+│ Sherlene302 │ Conrad619 │ Abington │
+│ Milan77 │ Conrad619 │ Abington │
+│ Frankie174 │ Seema671 │ Abington │
+│ Elane105 │ Josefina523 │ Abington │
+│ Lisha487 │ Shawanna357 │ Abington │
+│ Marleen824 │ Marcelino726 │ Abington │
+│ Sandi885 │ Lionel365 │ Abington │
+│ Morton637 │ Viva686 │ Abington │
+├──────────────┴───────────────┴──────────┤
+│ ? rows (>9999 rows, 20 shown) │
+└─────────────────────────────────────────┘
+```
+___
+___
+
+## Introduction to Subqueries and Nested Selects
+- Subqueries, also known as inner queries or nested queries, are SQL queries nested inside a larger query. They allow you to perform operations that usually require multiple steps in a single query, making your data retrieval process more efficient and concise. Nested selects are a type of subquery used specifically within the SELECT, FROM, or WHERE clauses to provide a dataset for the outer query to process.
+- Subqueries can return a single value, a single row, multiple rows, or a table. They are used for comparison, as a condition, or to provide a list of values for the outer query. The main distinction between correlated and non-correlated subqueries is that correlated subqueries reference column(s) from the outer query, thus running once for each row selected by the outer query, while non-correlated subqueries run independently of the outer query and can be run as standalone queries.
+
+### Types of subqueries
+* Single-Row Subqueries: Single-row subqueries return only one row and are used with single row comparison operators like =, >, <, >=, <=. They are often used in the WHERE clause to compare a column value against the result of the subquery.
+* Multi-Row Subqueries: Multi-row subqueries return more than one row and are used with operators like IN, ANY, ALL, which allow comparison against multiple values. They're useful for filtering based on a set of criteria returned by the subquery.
+* Correlated Subqueries: Correlated subqueries reference column(s) from the outer query, making them dependent on the outer query. They are executed once for each row processed by the outer query, often leading to performance considerations.
+
+___
+#### Use subqueries to find patients based on specific criteria.
+
+#### Find all patients who have been prescribed medication with a base cost higher than the average base cost of all medications.
+```python
+query = """SELECT p.FIRST, p.LAST
+FROM patients p
+WHERE p.Id IN ( SELECT m.PATIENT FROM medications m WHERE m.BASE_COST > (SELECT AVG(BASE_COST) FROM medications));
+"""
+conn.sql(query)
+```
+```python
+┌─────────────┬─────────────┐
+│ FIRST │ LAST │
+│ varchar │ varchar │
+├─────────────┼─────────────┤
+│ Dewitt635 │ Reichel38 │
+│ Anisa442 │ Purdy2 │
+│ Ross213 │ Mayert710 │
+│ Jim478 │ Mueller846 │
+│ Maria750 │ Schimmel440 │
+│ Ignacio928 │ Gorczany269 │
+│ Kip442 │ Zboncak558 │
+│ Jean712 │ Kuhlman484 │
+│ Mac103 │ Moen819 │
+│ Dianna917 │ Goldner995 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Cameron381 │ Bogan287 │
+│ Andrew29 │ Donnelly343 │
+│ Charis952 │ Littel644 │
+│ Lyndon118 │ Swift555 │
+│ Edward499 │ Zieme486 │
+│ Myron933 │ Ritchie586 │
+│ Yolonda722 │ Champlin946 │
+│ Alayna598 │ Kozey370 │
+│ Kristian973 │ Ledner144 │
+│ Sydney660 │ Zulauf375 │
+├─────────────┴─────────────┤
+│ ? rows 2 columns │
+└───────────────────────────┘
+```
+### Correlated Subqueries
+Learn how to use correlated subqueries to perform row-specific comparisons.
+Find patients whose healthcare expenses are higher than the average expenses in their county.
+```python
+query = """SELECT
+ p.FIRST,
+ p.LAST,
+ p.COUNTY,
+ p.HEALTHCARE_EXPENSES,
+ ROUND(AVG_EXPENSES.COUNTY_AVG_EXPENSES, 2) AS ROUNDED_AVG_HEALTHCARE_EXPENSES
+FROM
+ patients p
+INNER JOIN (
+ SELECT
+ COUNTY,
+ AVG(CAST(HEALTHCARE_EXPENSES AS DECIMAL(20,6))) AS COUNTY_AVG_EXPENSES
+ FROM
+ patients
+ GROUP BY
+ COUNTY
+) AS AVG_EXPENSES ON p.COUNTY = AVG_EXPENSES.COUNTY
+WHERE
+ p.HEALTHCARE_EXPENSES > (
+ SELECT AVG(p2.HEALTHCARE_EXPENSES)
+ FROM patients p2
+ WHERE p2.COUNTY = p.COUNTY
+ );
+"""
+conn.sql(query)
+```
+```python
+
+query = """SELECT
+ p.FIRST,
+ p.LAST,
+ p.COUNTY,
+ p.HEALTHCARE_EXPENSES,
+ ROUND(AVG_EXPENSES.COUNTY_AVG_EXPENSES, 2) AS ROUNDED_AVG_HEALTHCARE_EXPENSES
+FROM
+ patients p
+INNER JOIN (
+ SELECT
+… FROM patients p2
+ WHERE p2.COUNTY = p.COUNTY
+ );
+"""
+conn.sql(query)
+┌────────────┬───────────────┬───────────────────┬─────────────────────┬─────────────────────────────────┐
+│ FIRST │ LAST │ COUNTY │ HEALTHCARE_EXPENSES │ ROUNDED_AVG_HEALTHCARE_EXPENSES │
+│ varchar │ varchar │ varchar │ double │ double │
+├────────────┼───────────────┼───────────────────┼─────────────────────┼─────────────────────────────────┤
+│ Tammy740 │ Ernser583 │ Bristol County │ 1546025.67 │ 793296.73 │
+│ Iliana226 │ Schmeler639 │ Barnstable County │ 1407960.93 │ 949155.23 │
+│ Anthony633 │ Yundt842 │ Essex County │ 1575731.48 │ 814437.7 │
+│ Jim478 │ Mueller846 │ Bristol County │ 1112473.78 │ 793296.73 │
+│ Sina65 │ Howell947 │ Middlesex County │ 1479425.58 │ 841491.73 │
+│ Maria750 │ Schimmel440 │ Plymouth County │ 952814.55 │ 856778.1 │
+│ Lorenzo669 │ Vandervort697 │ Middlesex County │ 873001.47 │ 841491.73 │
+│ David908 │ Carter549 │ Worcester County │ 834576.02 │ 804752.74 │
+│ Noah480 │ O'Reilly797 │ Essex County │ 915110.6 │ 814437.7 │
+│ Kip442 │ Zboncak558 │ Middlesex County │ 1704103.7200000002 │ 841491.73 │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Tinisha932 │ Zulauf375 │ Bristol County │ 1035244.36 │ 793296.73 │
+│ Mario764 │ Estrada938 │ Suffolk County │ 1380401.5 │ 664340.79 │
+│ Glenn0 │ Schulist381 │ Barnstable County │ 1357123.8 │ 949155.23 │
+│ Porter490 │ Schulist381 │ Bristol County │ 1534216.57 │ 793296.73 │
+│ Darius626 │ Hackett68 │ Essex County │ 1715140.52 │ 814437.7 │
+│ Leonel449 │ Kunze215 │ Essex County │ 1492352.86 │ 814437.7 │
+│ Tyrell880 │ Kassulke119 │ Hampden County │ 1053520.21 │ 725636.71 │
+│ Arlen68 │ Gusikowski974 │ Worcester County │ 1363289.9 │ 804752.74 │
+│ Ashli227 │ Huel628 │ Worcester County │ 813730.02 │ 804752.74 │
+│ Loriann967 │ Mertz280 │ Norfolk County │ 1618397.57 │ 874351.83 │
+├────────────┴───────────────┴───────────────────┴─────────────────────┴─────────────────────────────────┤
+│ ? rows (>9999 rows, 20 shown) 5 columns │
+└────────────────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+Identify patients who have not been prescribed any medication.
+```python
+query = """
+SELECT p.FIRST, p.LAST
+FROM patients p
+WHERE NOT EXISTS (
+ SELECT 1 FROM medications m WHERE m.PATIENT = p.Id
+);
+"""
+
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT p.FIRST, p.LAST
+FROM patients p
+WHERE NOT EXISTS (
+ SELECT 1 FROM medications m WHERE m.PATIENT = p.Id
+);
+"""
+
+conn.sql(query)
+┌────────────┬───────────────┐
+│ FIRST │ LAST │
+│ varchar │ varchar │
+├────────────┼───────────────┤
+│ Iliana226 │ Schmeler639 │
+│ Toshiko149 │ Swaniawski813 │
+│ Heath320 │ Streich926 │
+│ Rana586 │ Langworth352 │
+│ Dorathy429 │ Yost751 │
+│ Deon400 │ Littel644 │
+│ Emerson869 │ D'Amore443 │
+│ Alec433 │ Sanford861 │
+│ Roselyn270 │ McLaughlin530 │
+│ Claudia969 │ Smith67 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Lou594 │ Gleason633 │
+│ Hank686 │ Russel238 │
+│ Earl438 │ Block661 │
+│ Mariano761 │ Heller342 │
+│ Sandra485 │ Fisher429 │
+│ Claire652 │ Murray856 │
+│ Doreen575 │ Graham902 │
+│ Fausto876 │ Kuhn96 │
+│ Vernon254 │ Gutmann970 │
+│ Ernest565 │ Block661 │
+├────────────┴───────────────┤
+│ ? rows 2 columns │
+└────────────────────────────┘
+```
+Find patients whose healthcare coverage is above the average of their respective city.
+```python
+query = """
+SELECT
+ p.FIRST,
+ p.LAST,
+ p.CITY,
+ ROUND(p.HEALTHCARE_COVERAGE,2) AS INDIVIDUAL_COVERAGE,
+ ROUND(c.AVG_COVERAGE,2) AS CITY_AVERAGE_COVERAGE
+FROM
+ patients p
+INNER JOIN (
+ SELECT
+ CITY,
+ AVG(HEALTHCARE_COVERAGE) AS AVG_COVERAGE
+ FROM
+ patients
+ GROUP BY
+ CITY
+) AS c ON p.CITY = c.CITY
+WHERE
+ p.HEALTHCARE_COVERAGE > (
+ SELECT AVG(HEALTHCARE_COVERAGE)
+ FROM patients
+ WHERE CITY = p.CITY
+ );
+"""
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT
+ p.FIRST,
+ p.LAST,
+ p.CITY,
+ ROUND(p.HEALTHCARE_COVERAGE,2) AS INDIVIDUAL_COVERAGE,
+ ROUND(c.AVG_COVERAGE,2) AS CITY_AVERAGE_COVERAGE
+FROM
+ patients p
+INNER JOIN (
+… SELECT AVG(HEALTHCARE_COVERAGE)
+ FROM patients
+ WHERE CITY = p.CITY
+ );
+"""
+conn.sql(query)
+┌──────────────┬────────────────┬───────────────┬─────────────────────┬───────────────────────┐
+│ FIRST │ LAST │ CITY │ INDIVIDUAL_COVERAGE │ CITY_AVERAGE_COVERAGE │
+│ varchar │ varchar │ varchar │ double │ double │
+├──────────────┼────────────────┼───────────────┼─────────────────────┼───────────────────────┤
+│ Anisa442 │ Purdy2 │ Methuen │ 34393.57 │ 12949.31 │
+│ Sina65 │ Howell947 │ Sudbury │ 15481.43 │ 15385.85 │
+│ Maria750 │ Schimmel440 │ Plymouth │ 14231.22 │ 10826.53 │
+│ Kip442 │ Zboncak558 │ Hopkinton │ 155478.44 │ 15439.17 │
+│ Hans694 │ Wilkinson796 │ Framingham │ 13729.73 │ 11988.99 │
+│ Jean712 │ Kuhlman484 │ Boxford │ 11421.48 │ 8660.53 │
+│ Lorette239 │ Abbott774 │ Dennis │ 78740.23 │ 14918.28 │
+│ Tobi258 │ Bernier607 │ Wilmington │ 13889.6 │ 9411.52 │
+│ Rosia390 │ Reichel38 │ Rochester │ 16234.29 │ 9736.74 │
+│ Florencia449 │ Mercado213 │ Wilbraham │ 11427.25 │ 10468.26 │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Jc393 │ Marks830 │ Brockton │ 16284.35 │ 12287.77 │
+│ Cedric746 │ Marvin195 │ Dedham │ 19806.72 │ 13840.99 │
+│ Milford485 │ Schmitt836 │ Hubbardston │ 10592.34 │ 8172.48 │
+│ Tegan755 │ Ruecker817 │ Waltham │ 30493.65 │ 14495.07 │
+│ Lindsay928 │ Lang846 │ Boston │ 11837.05 │ 10725.8 │
+│ Claudie965 │ Hermann103 │ Medford │ 15420.54 │ 12610.05 │
+│ Julie245 │ Nolan344 │ Essex │ 31903.53 │ 31787.13 │
+│ Mui729 │ Stoltenberg489 │ Wakefield │ 21528.91 │ 13699.32 │
+│ Oliver401 │ Lesch175 │ Yarmouth │ 27442.87 │ 19907.83 │
+│ Mi492 │ Gusikowski974 │ North Andover │ 18481.09 │ 17677.13 │
+├──────────────┴────────────────┴───────────────┴─────────────────────┴───────────────────────┤
+│ ? rows (>9999 rows, 20 shown) 5 columns │
+└─────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+## FUNCTIONS IN SQL
+___
+### String Functions
+### CONCAT: Concatenates two or more strings.
+```python
+query = """SELECT CONCAT(FIRST, ' ', LAST) AS FullName FROM patients LIMIT 10;
+;"""
+conn.sql(query)
+```
+```python
+┌───────────────────────────┐
+│ FullName │
+│ varchar │
+├───────────────────────────┤
+│ Jacinto644 Kris249 │
+│ Alva958 Krajcik437 │
+│ Jimmie93 Harris789 │
+│ Gregorio366 Auer97 │
+│ Karyn217 Mueller846 │
+│ Jayson808 Fadel536 │
+│ José Eduardo181 Gómez206 │
+│ Milo271 Feil794 │
+│ Karyn217 Metz686 │
+│ Jeffrey461 Greenfelder433 │
+├───────────────────────────┤
+│ 10 rows │
+└───────────────────────────┘
+```
+### DATALENGTH: Returns the number of bytes used to represent any expre
+```python
+query = """
+SELECT FIRST, LENGTH(FIRST) AS LengthInBytes FROM patients LIMIT 10;
+"""
+
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT FIRST, LENGTH(FIRST) AS LengthInBytes FROM patients LIMIT 10;
+"""
+
+conn.sql(query)
+┌─────────────────┬───────────────┐
+│ FIRST │ LengthInBytes │
+│ varchar │ int64 │
+├─────────────────┼───────────────┤
+│ Jacinto644 │ 10 │
+│ Alva958 │ 7 │
+│ Jimmie93 │ 8 │
+│ Gregorio366 │ 11 │
+│ Karyn217 │ 8 │
+│ Jayson808 │ 9 │
+│ José Eduardo181 │ 15 │
+│ Milo271 │ 7 │
+│ Karyn217 │ 8 │
+│ Jeffrey461 │ 10 │
+├─────────────────┴───────────────┤
+│ 10 rows 2 columns │
+└─────────────────────────────────┘
+```
+### LEFT: Returns the left part of a character string with the specified number of characters.
+```python
+query = """SELECT LEFT(FIRST, 5) AS Initial FROM patients LIMIT 10;"""
+conn.sql(query)
+```
+```python
+
+query = """SELECT LEFT(FIRST, 5) AS Initial FROM patients LIMIT 10;"""
+conn.sql(query)
+┌─────────┐
+│ Initial │
+│ varchar │
+├─────────┤
+│ Jacin │
+│ Alva9 │
+│ Jimmi │
+│ Grego │
+│ Karyn │
+│ Jayso │
+│ José │
+│ Milo2 │
+│ Karyn │
+│ Jeffr │
+├─────────┤
+│ 10 rows │
+└─────────┘
+```
+### LOWER: Converts all characters in the specified string to lowercase.
+### UPPER: Convert to uppercase
+```python
+query = """
+SELECT LOWER(FIRST) AS LowercaseFirstName, UPPER(LAST) AS UppercaseLastName FROM patients LIMIT 10;
+"""
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT LOWER(FIRST) AS LowercaseFirstName, UPPER(LAST) AS UppercaseLastName FROM patients LIMIT 10;
+"""
+conn.sql(query)
+┌────────────────────┬───────────────────┐
+│ LowercaseFirstName │ UppercaseLastName │
+│ varchar │ varchar │
+├────────────────────┼───────────────────┤
+│ jacinto644 │ KRIS249 │
+│ alva958 │ KRAJCIK437 │
+│ jimmie93 │ HARRIS789 │
+│ gregorio366 │ AUER97 │
+│ karyn217 │ MUELLER846 │
+│ jayson808 │ FADEL536 │
+│ josé eduardo181 │ GÓMEZ206 │
+│ milo271 │ FEIL794 │
+│ karyn217 │ METZ686 │
+│ jeffrey461 │ GREENFELDER433 │
+├────────────────────┴───────────────────┤
+│ 10 rows 2 columns │
+└────────────────────────────────────────┘
+```
+LTRIM: Removes leading spaces from a string.
+```python
+query = """SELECT LTRIM(ADDRESS) AS TrimmedAddress FROM patients LIMIT 10;"""
+
+conn.sql(query)
+
+```
+```python
+
+query = """SELECT LTRIM(ADDRESS) AS TrimmedAddress FROM patients LIMIT 10;"""
+
+conn.sql(query)
+
+┌────────────────────────────────┐
+│ TrimmedAddress │
+│ varchar │
+├────────────────────────────────┤
+│ 888 Hickle Ferry Suite 38 │
+│ 1048 Skiles Trailer │
+│ 201 Mitchell Lodge Unit 67 │
+│ 1050 Lindgren Extension Apt 38 │
+│ 570 Abshire Forge Suite 32 │
+│ 1056 Harris Lane Suite 70 │
+│ 427 Balistreri Way Unit 19 │
+│ 422 Farrell Path Unit 69 │
+│ 181 Feest Passage Suite 64 │
+│ 428 Wiza Glen Unit 91 │
+├────────────────────────────────┤
+│ 10 rows │
+└────────────────────────────────┘
+```
+REPLACE: Replaces all occurrences of a specified string value with another string value.
+```python
+query = """SELECT FIRST, LAST,
+ ADDRESS AS LongAddress,
+ REPLACE(ADDRESS, 'Street', 'St') AS ShortAddress
+FROM
+ patients
+WHERE
+ ADDRESS LIKE '%Street%' LIMIT 10;
+"""
+
+conn.sql(query)
+```
+```python
+┌────────────┬───────────────┬───────────────────────────┬───────────────────────┐
+│ FIRST │ LAST │ LongAddress │ ShortAddress │
+│ varchar │ varchar │ varchar │ varchar │
+├────────────┼───────────────┼───────────────────────────┼───────────────────────┤
+│ Lorrie905 │ Leannon79 │ 813 Casper Street │ 813 Casper St │
+│ Asa127 │ Block661 │ 140 Rohan Street Suite 50 │ 140 Rohan St Suite 50 │
+│ Logan497 │ Brekke496 │ 1081 Orn Street │ 1081 Orn St │
+│ Cletus494 │ Strosin214 │ 1019 Haley Street │ 1019 Haley St │
+│ Cortney940 │ Stehr398 │ 458 Streich Street │ 458 Streich St │
+│ Latoyia537 │ Gaylord332 │ 200 Heaney Street │ 200 Heaney St │
+│ Clark193 │ Hilll811 │ 202 Tromp Street Suite 0 │ 202 Tromp St Suite 0 │
+│ Jc393 │ Bosco882 │ 792 Walsh Street │ 792 Walsh St │
+│ Noel608 │ Swaniawski813 │ 155 Walter Street │ 155 Walter St │
+│ Sari509 │ Hoppe518 │ 550 Lang Street Suite 65 │ 550 Lang St Suite 65 │
+├────────────┴───────────────┴───────────────────────────┴───────────────────────┤
+│ 10 rows 4 columns │
+└────────────────────────────────────────────────────────────────────────────────┘
+```
+RIGHT: Returns the right part of a character string with the specified number of characters.
+```python
+query = """SELECT FIRST, RIGHT(FIRST, 6) AS LastSixChars FROM patients LIMIT 10;
+"""
+
+conn.sql(query)
+```
+```python
+┌─────────────────┬──────────────┐
+│ FIRST │ LastSixChars │
+│ varchar │ varchar │
+├─────────────────┼──────────────┤
+│ Jacinto644 │ nto644 │
+│ Alva958 │ lva958 │
+│ Jimmie93 │ mmie93 │
+│ Gregorio366 │ rio366 │
+│ Karyn217 │ ryn217 │
+│ Jayson808 │ son808 │
+│ José Eduardo181 │ rdo181 │
+│ Milo271 │ ilo271 │
+│ Karyn217 │ ryn217 │
+│ Jeffrey461 │ rey461 │
+├─────────────────┴──────────────┤
+│ 10 rows 2 columns │
+└────────────────────────────────┘
+```
+SUBSTRING: Returns part of a character, binary, text, or image expression in SQL Server.
+```python
+query = """SELECT FIRST, SUBSTRING(FIRST, 1, 4) AS FirstFourChars FROM patients LIMIT 5;
+"""
+
+conn.sql(query)
+```
+```python
+┌─────────────┬────────────────┐
+│ FIRST │ FirstFourChars │
+│ varchar │ varchar │
+├─────────────┼────────────────┤
+│ Jacinto644 │ Jaci │
+│ Alva958 │ Alva │
+│ Jimmie93 │ Jimm │
+│ Gregorio366 │ Greg │
+│ Karyn217 │ Kary │
+└─────────────┴────────────────┘
+```
+REGEXP_REPLACE can use regular expressions to strip numeric characters:
+```python
+query = """SELECT
+FIRST, REGEXP_REPLACE(FIRST, '[0-9]', '', 'g') AS FirstNameStripped, LAST, REGEXP_REPLACE(LAST, '[0-9]', '', 'g') AS LastNameStripped
+FROM patients;
+"""
+
+conn.sql(query)
+```
+```python
+
+query = """SELECT
+FIRST, REGEXP_REPLACE(FIRST, '[0-9]', '', 'g') AS FirstNameStripped, LAST, REGEXP_REPLACE(LAST, '[0-9]', '', 'g') AS LastNameStripped
+FROM patients;
+"""
+
+conn.sql(query)
+┌─────────────────┬───────────────────┬────────────────┬──────────────────┐
+│ FIRST │ FirstNameStripped │ LAST │ LastNameStripped │
+│ varchar │ varchar │ varchar │ varchar │
+├─────────────────┼───────────────────┼────────────────┼──────────────────┤
+│ Jacinto644 │ Jacinto │ Kris249 │ Kris │
+│ Alva958 │ Alva │ Krajcik437 │ Krajcik │
+│ Jimmie93 │ Jimmie │ Harris789 │ Harris │
+│ Gregorio366 │ Gregorio │ Auer97 │ Auer │
+│ Karyn217 │ Karyn │ Mueller846 │ Mueller │
+│ Jayson808 │ Jayson │ Fadel536 │ Fadel │
+│ José Eduardo181 │ José Eduardo │ Gómez206 │ Gómez │
+│ Milo271 │ Milo │ Feil794 │ Feil │
+│ Karyn217 │ Karyn │ Metz686 │ Metz │
+│ Jeffrey461 │ Jeffrey │ Greenfelder433 │ Greenfelder │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Raymond398 │ Raymond │ Kuvalis369 │ Kuvalis │
+│ Gearldine455 │ Gearldine │ Boyer713 │ Boyer │
+│ Nichol11 │ Nichol │ Gleichner915 │ Gleichner │
+│ Louvenia131 │ Louvenia │ Marks830 │ Marks │
+│ Raymon366 │ Raymon │ Beer512 │ Beer │
+│ Camelia346 │ Camelia │ Stamm704 │ Stamm │
+│ William805 │ William │ Pacocha935 │ Pacocha │
+│ Guillermo498 │ Guillermo │ Téllez750 │ Téllez │
+│ Milton509 │ Milton │ Bailey598 │ Bailey │
+│ Cecilia788 │ Cecilia │ Wisozk929 │ Wisozk │
+├─────────────────┴───────────────────┴────────────────┴──────────────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└─────────────────────────────────────────────────────────────────────────┘
+```
+___
+___
+
+### Creating a database and adding data to tables
+
+### CREATE, ALTER, DROP, INSERT commands
+___
+First lets have a in-memory database
+```python
+newdb = duckdb.connect()
+```
+CREATE TABLE: Create a books table to mimic a library catalog system.
+```python
+query = """CREATE TABLE books (
+ book_id INTEGER PRIMARY KEY,
+ title VARCHAR,
+ author VARCHAR,
+ published_year INTEGER
+);
+"""
+
+newdb.sql(query)
+```
+The CREATE TABLE statement defines the schema for this table:
+This statement creates a table with columns for the book ID, title, author, and the year of publication. The book_id column is designated as the primary key, ensuring that each record in the table is unique.
+___
+check
+```python
+newdb.sql("SHOW TABLES;")
+```
+```python
+┌─────────┐
+│ name │
+│ varchar │
+├─────────┤
+│ books │
+└─────────┘
+```
+check data
+```python
+newdb.sql("DESCRIBE books;")
+```
+```python
+
+newdb.sql("DESCRIBE books;")
+┌────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ book_id │ INTEGER │ NO │ PRI │ NULL │ NULL │
+│ title │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ author │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ published_year │ INTEGER │ YES │ NULL │ NULL │ NULL │
+└────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┘
+```
+Alter Table: After creating the table, you might need to add more information. Use the ALTER TABLE statement to add a new column:
+```python
+query = """ALTER TABLE books ADD COLUMN genre VARCHAR;
+"""
+newdb.sql(query)
+# This adds a genre column to the books table, allowing you to categorize books.
+```
+```python
+newdb.sql("DESCRIBE books;")
+```
+```python
+
+newdb.sql("DESCRIBE books;")
+┌────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ book_id │ INTEGER │ NO │ PRI │ NULL │ NULL │
+│ title │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ author │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ published_year │ INTEGER │ YES │ NULL │ NULL │ NULL │
+│ genre │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+└────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┘
+```
+let's add another table which we will DROP later;
+```python
+query = """CREATE TABLE authors (
+ author_id INTEGER PRIMARY KEY,
+ firstname VARCHAR,
+ lastname VARCHAR,
+ birth_year INTEGER
+);
+"""
+
+newdb.sql(query)
+```
+
+```python
+newdb.sql("SHOW TABLES;")
+```
+Now let's DROP the author table
+```python
+query = """DROP TABLE IF EXISTS authors;
+"""
+
+newdb.sql(query)
+```
+
+```python
+newdb.sql("SHOW TABLES;")
+```
+### Let's add some data now
+### INSERT To populate the books table with data. This inserts 10 records into the books table, providing a diverse set of examples.
+```python
+query = """INSERT INTO books (book_id, title, author, published_year, genre) VALUES
+(1, '1984', 'George Orwell', 1949, 'Dystopian'),
+(2, 'The Great Gatsby', 'F. Scott Fitzgerald', 1925, 'Classic'),
+(3, 'To Kill a Mockingbird', 'Harper Lee', 1960, 'Classic'),
+(4, 'Pride and Prejudice', 'Jane Austen', 1813, 'Romance'),
+(5, 'The Hobbit', 'J.R.R. Tolkien', 1937, 'Fantasy'),
+(6, 'The Catcher in the Rye', 'J.D. Salinger', 1951, 'Literary Fiction'),
+(7, 'Harry Potter and the Sorcerer’s Stone', 'J.K. Rowling', 1997, 'Fantasy'),
+(8, 'The Da Vinci Code', 'Dan Brown', 2003, 'Thriller'),
+(9, 'Sapiens: A Brief History of Humankind', 'Yuval Noah Harari', 2011, 'Non-fiction'),
+(10, 'Beloved', 'Toni Morrison', 1987, 'Historical Fiction');"""
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT title AS "Book Title", author AS "Author Name", Genre FROM books;
+"""
+
+newdb.sql(query)
+```
+
+```python
+┌───────────────────────────────────────┬─────────────────────┬────────────────────┐
+│ Book Title │ Author Name │ genre │
+│ varchar │ varchar │ varchar │
+├───────────────────────────────────────┼─────────────────────┼────────────────────┤
+│ 1984 │ George Orwell │ Dystopian │
+│ The Great Gatsby │ F. Scott Fitzgerald │ Classic │
+│ To Kill a Mockingbird │ Harper Lee │ Classic │
+│ Pride and Prejudice │ Jane Austen │ Romance │
+│ The Hobbit │ J.R.R. Tolkien │ Fantasy │
+│ The Catcher in the Rye │ J.D. Salinger │ Literary Fiction │
+│ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ Fantasy │
+│ The Da Vinci Code │ Dan Brown │ Thriller │
+│ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ Non-fiction │
+│ Beloved │ Toni Morrison │ Historical Fiction │
+├───────────────────────────────────────┴─────────────────────┴────────────────────┤
+│ 10 rows 3 columns │
+└──────────────────────────────────────────────────────────────────────────────────┘
+```
+### Updating Data: UPDATE
+This changes the genre of the book titled '1984' to 'Science Fiction'.
+```python
+query = """UPDATE books SET genre = 'Science Fiction' WHERE title = '1984';
+"""
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT title AS "Book Title", author AS "Author Name", Genre FROM books;
+"""
+
+newdb.sql(query)
+```
+
+```python
+┌───────────────────────────────────────┬─────────────────────┬────────────────────┐
+│ Book Title │ Author Name │ genre │
+│ varchar │ varchar │ varchar │
+├───────────────────────────────────────┼─────────────────────┼────────────────────┤
+│ 1984 │ George Orwell │ Science Fiction │
+│ The Great Gatsby │ F. Scott Fitzgerald │ Classic │
+│ To Kill a Mockingbird │ Harper Lee │ Classic │
+│ Pride and Prejudice │ Jane Austen │ Romance │
+│ The Hobbit │ J.R.R. Tolkien │ Fantasy │
+│ The Catcher in the Rye │ J.D. Salinger │ Literary Fiction │
+│ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ Fantasy │
+│ The Da Vinci Code │ Dan Brown │ Thriller │
+│ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ Non-fiction │
+│ Beloved │ Toni Morrison │ Historical Fiction │
+├───────────────────────────────────────┴─────────────────────┴────────────────────┤
+│ 10 rows 3 columns │
+└──────────────────────────────────────────────────────────────────────────────────┘
+```
+## DELETE
+e.g. To remove a book from the catalog
+
+```python
+query = """DELETE FROM books WHERE title = 'The Great Gatsby';"""
+newdb.sql(query)
+```
+
+```python
+query = """SELECT title AS "Book Title", author AS "Author Name", Genre FROM books;
+"""
+
+newdb.sql(query)
+```
+
+```python
+┌───────────────────────────────────────┬───────────────────┬────────────────────┐
+│ Book Title │ Author Name │ genre │
+│ varchar │ varchar │ varchar │
+├───────────────────────────────────────┼───────────────────┼────────────────────┤
+│ 1984 │ George Orwell │ Science Fiction │
+│ To Kill a Mockingbird │ Harper Lee │ Classic │
+│ Pride and Prejudice │ Jane Austen │ Romance │
+│ The Hobbit │ J.R.R. Tolkien │ Fantasy │
+│ The Catcher in the Rye │ J.D. Salinger │ Literary Fiction │
+│ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ Fantasy │
+│ The Da Vinci Code │ Dan Brown │ Thriller │
+│ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ Non-fiction │
+│ Beloved │ Toni Morrison │ Historical Fiction │
+└───────────────────────────────────────┴───────────────────┴────────────────────┘
+```
+### TRUNCATE
+- The TRUNCATE TABLE statement is used to delete all rows in a table without logging the individual row deletions.
+- This is faster than using DELETE without a WHERE clause because TRUNCATE immediately deallocates data pages used by the table.
+- Use TRUNCATE when you want to quickly remove all records from a table but keep the table structure for future use.
+- Unlike DELETE, TRUNCATE does not generate a large number of transaction logs, making it more efficient for completely clearing a table.
+
+```python
+query = """TRUNCATE TABLE books;"""
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT * FROM books;
+"""
+
+newdb.sql(query)
+```
+#### Adding one more table; named 'authors'
+```python
+query = """CREATE TABLE authors (
+ author_id INTEGER PRIMARY KEY,
+ name VARCHAR,
+ birth_year INTEGER,
+ nationality VARCHAR
+);
+"""
+
+newdb.sql(query)
+```
+Add information the authors table
+```python
+query = """INSERT INTO authors (author_id, name, birth_year, nationality) VALUES
+(1, 'George Orwell', 1903, 'British'),
+(2, 'F. Scott Fitzgerald', 1896, 'American'),
+(3, 'Harper Lee', 1926, 'American'),
+(4, 'Jane Austen', 1775, 'British'),
+(5, 'J.R.R. Tolkien', 1892, 'British'),
+(6, 'J.D. Salinger', 1919, 'American'),
+(7, 'J.K. Rowling', 1965, 'British'),
+(8, 'Dan Brown', 1964, 'American'),
+(9, 'Yuval Noah Harari', 1976, 'Israeli'),
+(10, 'Toni Morrison', 1931, 'American');
+"""
+
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT * FROM books;
+"""
+
+newdb.sql(query)
+```
+```python
+┌─────────┬───────────────────────────────────────┬─────────────────────┬────────────────┬────────────────────┐
+│ book_id │ title │ author │ published_year │ genre │
+│ int64 │ varchar │ varchar │ int64 │ varchar │
+├─────────┼───────────────────────────────────────┼─────────────────────┼────────────────┼────────────────────┤
+│ 1 │ 1984 │ George Orwell │ 1949 │ Dystopian │
+│ 2 │ The Great Gatsby │ F. Scott Fitzgerald │ 1925 │ Classic │
+│ 3 │ To Kill a Mockingbird │ Harper Lee │ 1960 │ Classic │
+│ 4 │ Pride and Prejudice │ Jane Austen │ 1813 │ Romance │
+│ 5 │ The Hobbit │ J.R.R. Tolkien │ 1937 │ Fantasy │
+│ 6 │ The Catcher in the Rye │ J.D. Salinger │ 1951 │ Literary Fiction │
+│ 7 │ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ 1997 │ Fantasy │
+│ 8 │ The Da Vinci Code │ Dan Brown │ 2003 │ Thriller │
+│ 9 │ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ 2011 │ Non-fiction │
+│ 10 │ Beloved │ Toni Morrison │ 1987 │ Historical Fiction │
+├─────────┴───────────────────────────────────────┴─────────────────────┴────────────────┴────────────────────┤
+│ 10 rows 5 columns │
+└─────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+
+```python
+query = """SELECT * FROM authors;
+"""
+
+newdb.sql(query)
+```
+
+```python
+newdb.sql(query)
+┌───────────┬─────────────────────┬────────────┬─────────────┐
+│ author_id │ name │ birth_year │ nationality │
+│ int32 │ varchar │ int32 │ varchar │
+├───────────┼─────────────────────┼────────────┼─────────────┤
+│ 1 │ George Orwell │ 1903 │ British │
+│ 2 │ F. Scott Fitzgerald │ 1896 │ American │
+│ 3 │ Harper Lee │ 1926 │ American │
+│ 4 │ Jane Austen │ 1775 │ British │
+│ 5 │ J.R.R. Tolkien │ 1892 │ British │
+│ 6 │ J.D. Salinger │ 1919 │ American │
+│ 7 │ J.K. Rowling │ 1965 │ British │
+│ 8 │ Dan Brown │ 1964 │ American │
+│ 9 │ Yuval Noah Harari │ 1976 │ Israeli │
+│ 10 │ Toni Morrison │ 1931 │ American │
+├───────────┴─────────────────────┴────────────┴─────────────┤
+│ 10 rows 4 columns │
+└────────────────────────────────────────────────────────────┘
+```
+## EXPORTING Data to csv from SQL
+```python
+query = """EXPORT DATABASE './' (FORMAT CSV, DELIMITER ',');"""
+
+newdb.sql(query)
+```
+## Thank You!
diff --git a/11_sql_duckdb/index.html b/11_sql_duckdb/index.html
new file mode 100644
index 000000000..2a394295c
--- /dev/null
+++ b/11_sql_duckdb/index.html
@@ -0,0 +1,3487 @@
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+* An in-memory, columnar database management system optimized for analytical queries."
+* Open-source and designed for simplicity, speed, and efficiency in processing analytical workloads."
+* Supports SQL standards for easy integration with existing tools and workflows.
+___
+## Install DuckDB
+### With Python set up, you can now install DuckDB using pip. In your terminal or command line, execute:
+```console
+pip install duckdb
+```
+___
+
+
+
+___
+
+### To ensure DuckDB is installed correctly, launch your jupyter Notebook and try importing the DuckDB module:
+
+```python
+pip install duckdb
+```
+### If no errors occur, DuckDB is successfully installed and ready to use.
+___
+### In your Jupyter Notebook, import duckdb by running
+```python
+import duckdb
+```
+### For this tutorial, we will be using a pre-configured Duckdb Database.
+### Let's download the database file:
+```python
+!wget --content-disposition https://arizona.box.com/shared/static/uozg0z86rtdjupwpc7i971xwzuzhp42o.duckdb
+```
+
+### Connect to the database using:
+```python
+conn = duckdb.connect(database='/content/my_database.duckdb', read_only=True)
+```
+### Viewing which tables are available inside the database
+
+```python
+conn.sql("SHOW TABLES;")
+```
+
+```python
+┌───────────────────┐
+│ name │
+│ varchar │
+├───────────────────┤
+│ allergies │
+│ careplans │
+│ conditions │
+│ devices │
+│ encounters │
+│ imaging_studies │
+│ immunizations │
+│ medications │
+│ observations │
+│ organizations │
+│ patients │
+│ payer_transitions │
+│ payers │
+│ procedures │
+│ providers │
+│ supplies │
+├───────────────────┤
+│ 16 rows │
+└───────────────────┘
+```
+### Before running any query, we need to know the columns inside particular tables
+
+```python
+conn.sql("DESCRIBE patients;")
+```
+```python
+┌─────────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├─────────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ Id │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ BIRTHDATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ DEATHDATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ SSN │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ DRIVERS │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PASSPORT │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PREFIX │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ FIRST │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ LAST │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ SUFFIX │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ · │ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │ · │
+│ BIRTHPLACE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ADDRESS │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ CITY │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ STATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ COUNTY │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ZIP │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ LAT │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ LON │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ HEALTHCARE_EXPENSES │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ HEALTHCARE_COVERAGE │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+├─────────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┤
+│ 25 rows (20 shown) 6 columns │
+└─────────────────────────────────────────────────────────────────────────┘
+```
+```python
+conn.sql("DESCRIBE medications;")
+```
+```python
+
+conn.sql("DESCRIBE medications;")
+┌───────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├───────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ START │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ STOP │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PATIENT │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PAYER │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ENCOUNTER │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ CODE │ BIGINT │ YES │ NULL │ NULL │ NULL │
+│ DESCRIPTION │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ BASE_COST │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ PAYER_COVERAGE │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ DISPENSES │ BIGINT │ YES │ NULL │ NULL │ NULL │
+│ TOTALCOST │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ REASONCODE │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+│ REASONDESCRIPTION │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+├───────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┤
+│ 13 rows 6 columns │
+└───────────────────────────────────────────────────────────────────────┘
+```
+```python
+conn.sql("DESCRIBE immunizations;")
+```
+```python
+┌─────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├─────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ DATE │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ PATIENT │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ ENCOUNTER │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ CODE │ BIGINT │ YES │ NULL │ NULL │ NULL │
+│ DESCRIPTION │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ BASE_COST │ DOUBLE │ YES │ NULL │ NULL │ NULL │
+└─────────────┴─────────────┴─────────┴─────────┴─────────┴───────┘
+```
+___
+___
+### Querying Data
+#### **SELECT** - The SELECT statement selects data from a database.
+
+E.g.: Query the patients table and display patient first name, last name, gender, and the city they live in.
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients;")
+```
+```python
+┌─────────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├─────────────────┼────────────────┼─────────┼─────────────┤
+│ Jacinto644 │ Kris249 │ M │ Springfield │
+│ Alva958 │ Krajcik437 │ F │ Walpole │
+│ Jimmie93 │ Harris789 │ F │ Pembroke │
+│ Gregorio366 │ Auer97 │ M │ Boston │
+│ Karyn217 │ Mueller846 │ F │ Colrain │
+│ Jayson808 │ Fadel536 │ M │ Chicopee │
+│ José Eduardo181 │ Gómez206 │ M │ Chicopee │
+│ Milo271 │ Feil794 │ M │ Somerville │
+│ Karyn217 │ Metz686 │ F │ Medfield │
+│ Jeffrey461 │ Greenfelder433 │ M │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Raymond398 │ Kuvalis369 │ M │ Hingham │
+│ Gearldine455 │ Boyer713 │ F │ Boston │
+│ Nichol11 │ Gleichner915 │ F │ Lynn │
+│ Louvenia131 │ Marks830 │ F │ Springfield │
+│ Raymon366 │ Beer512 │ M │ Springfield │
+│ Camelia346 │ Stamm704 │ F │ Dedham │
+│ William805 │ Pacocha935 │ M │ Brockton │
+│ Guillermo498 │ Téllez750 │ M │ Somerville │
+│ Milton509 │ Bailey598 │ M │ Hull │
+│ Cecilia788 │ Wisozk929 │ F │ Worcester │
+├─────────────────┴────────────────┴─────────┴─────────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────┘
+```
+### Count the number of rows in patients table
+### This query will count the total number of rows in the patients table, effectively giving you the total number of patients.
+
+```python
+conn.sql("SELECT COUNT(*) FROM patients;")
+```
+```python
+┌──────────────┐
+│ count_star() │
+│ int64 │
+├──────────────┤
+│ 124150 │
+└──────────────┘
+```
+### **COUNT** and **DISTINCT**
+If you want to count distinct values of a specific column, for example, distinct cities, you can modify the query as follows:
+```python
+conn.sql("SELECT COUNT(DISTINCT CITY) FROM patients;")
+```
+```python
+┌──────────────────────┐
+│ count(DISTINCT CITY) │
+│ int64 │
+├──────────────────────┤
+│ 351 │
+└──────────────────────┘
+```
+### Filtering data based on a condition
+The WHERE clause filters records that fulfill a specified condition.
+e.g: Patients in City = 'Springfield';
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY, PASSPORT FROM patients WHERE city = 'Springfield';")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┬────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │ PASSPORT │
+│ varchar │ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┼────────────┤
+│ Jacinto644 │ Kris249 │ M │ Springfield │ NULL │
+│ Jeffrey461 │ Greenfelder433 │ M │ Springfield │ NULL │
+│ Sabina296 │ Flatley871 │ F │ Springfield │ X85058581X │
+│ Theodora872 │ Johnson679 │ F │ Springfield │ X21164602X │
+│ Lavera253 │ Anderson154 │ F │ Springfield │ X83686992X │
+│ Golden321 │ Pollich983 │ F │ Springfield │ NULL │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │ X58134116X │
+│ Fausto876 │ Bechtelar572 │ M │ Springfield │ NULL │
+│ Talisha682 │ Brakus656 │ F │ Springfield │ X53645004X │
+│ Golden321 │ Durgan499 │ F │ Springfield │ X49016634X │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Carla633 │ Terán294 │ F │ Springfield │ X50212869X │
+│ Ángela136 │ Muñiz642 │ F │ Springfield │ NULL │
+│ Marceline716 │ Kuhlman484 │ F │ Springfield │ X70432308X │
+│ Ana María762 │ Cotto891 │ F │ Springfield │ X4604857X │
+│ Josefina523 │ Vanegas191 │ F │ Springfield │ X85396222X │
+│ Ester635 │ Sevilla788 │ F │ Springfield │ X50601786X │
+│ Tomás404 │ Galarza986 │ M │ Springfield │ X76790663X │
+│ Arturo47 │ Delafuente833 │ M │ Springfield │ X45087804X │
+│ Gilberto712 │ Martínez540 │ M │ Springfield │ X89729075X │
+│ Juanita470 │ Connelly992 │ F │ Springfield │ X71156217X │
+├──────────────┴────────────────┴─────────┴─────────────┴────────────┤
+│ 2814 rows (20 shown) 5 columns │
+└────────────────────────────────────────────────────────────────────┘
+```
+### Multiple conditions in Selection
+e.g 1: Female Patients in City = 'Springfield';
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE city = 'Springfield' AND gender = 'F';")
+```
+```python
+
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE city = 'Springfield' AND gender = 'F';")
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │
+│ Theodora872 │ Johnson679 │ F │ Springfield │
+│ Lavera253 │ Anderson154 │ F │ Springfield │
+│ Golden321 │ Pollich983 │ F │ Springfield │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Golden321 │ Durgan499 │ F │ Springfield │
+│ Jerrie417 │ Gislason620 │ F │ Springfield │
+│ Venus149 │ Hodkiewicz467 │ F │ Springfield │
+│ Refugia211 │ Wintheiser220 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Tisha655 │ Renner328 │ F │ Springfield │
+│ Mona85 │ Senger904 │ F │ Springfield │
+│ Isabel214 │ Camacho176 │ F │ Springfield │
+│ Carla633 │ Terán294 │ F │ Springfield │
+│ Ángela136 │ Muñiz642 │ F │ Springfield │
+│ Marceline716 │ Kuhlman484 │ F │ Springfield │
+│ Ana María762 │ Cotto891 │ F │ Springfield │
+│ Josefina523 │ Vanegas191 │ F │ Springfield │
+│ Ester635 │ Sevilla788 │ F │ Springfield │
+│ Juanita470 │ Connelly992 │ F │ Springfield │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 1351 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+### Nested selection
+e.g. Female patients from City Springfield or Boston
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE gender = 'F' AND (city = 'Springfield' OR city = 'Boston');")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │
+│ Theodora872 │ Johnson679 │ F │ Springfield │
+│ Lavera253 │ Anderson154 │ F │ Springfield │
+│ Golden321 │ Pollich983 │ F │ Springfield │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Golden321 │ Durgan499 │ F │ Springfield │
+│ Jerrie417 │ Gislason620 │ F │ Springfield │
+│ Venus149 │ Hodkiewicz467 │ F │ Springfield │
+│ Refugia211 │ Wintheiser220 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Freida957 │ Hand679 │ F │ Boston │
+│ Clora637 │ Rempel203 │ F │ Boston │
+│ Bonny428 │ Turner526 │ F │ Boston │
+│ Tien590 │ Gaylord332 │ F │ Boston │
+│ Arlyne429 │ Deckow585 │ F │ Boston │
+│ Doreen575 │ Johnson679 │ F │ Boston │
+│ Kym935 │ Hayes766 │ F │ Boston │
+│ Julia241 │ Nevárez403 │ F │ Boston │
+│ Ja391 │ Murray856 │ F │ Boston │
+│ Creola518 │ Spinka232 │ F │ Boston │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 7191 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+#### Alternative
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE gender = 'F' AND (city IN ('Springfield','Boston'));")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │
+│ Theodora872 │ Johnson679 │ F │ Springfield │
+│ Lavera253 │ Anderson154 │ F │ Springfield │
+│ Golden321 │ Pollich983 │ F │ Springfield │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Golden321 │ Durgan499 │ F │ Springfield │
+│ Jerrie417 │ Gislason620 │ F │ Springfield │
+│ Venus149 │ Hodkiewicz467 │ F │ Springfield │
+│ Refugia211 │ Wintheiser220 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Freida957 │ Hand679 │ F │ Boston │
+│ Clora637 │ Rempel203 │ F │ Boston │
+│ Bonny428 │ Turner526 │ F │ Boston │
+│ Tien590 │ Gaylord332 │ F │ Boston │
+│ Arlyne429 │ Deckow585 │ F │ Boston │
+│ Doreen575 │ Johnson679 │ F │ Boston │
+│ Kym935 │ Hayes766 │ F │ Boston │
+│ Julia241 │ Nevárez403 │ F │ Boston │
+│ Ja391 │ Murray856 │ F │ Boston │
+│ Creola518 │ Spinka232 │ F │ Boston │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 7191 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+### Filter on missing data
+* Filtering on missing data is crucial for maintaining data integrity and ensuring accurate analysis by identifying and handling incomplete records effectively.
+* Retrieve the first name, last name, gender, and city for all patients who are from Springfield and have a passport number recorded (i.e., the passport field is not empty).
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY, PASSPORT FROM patients WHERE city = 'Springfield' AND PASSPORT IS NOT NULL;")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┬────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │ PASSPORT │
+│ varchar │ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┼────────────┤
+│ Sabina296 │ Flatley871 │ F │ Springfield │ X85058581X │
+│ Theodora872 │ Johnson679 │ F │ Springfield │ X21164602X │
+│ Lavera253 │ Anderson154 │ F │ Springfield │ X83686992X │
+│ Georgiann138 │ Greenfelder433 │ F │ Springfield │ X58134116X │
+│ Talisha682 │ Brakus656 │ F │ Springfield │ X53645004X │
+│ Golden321 │ Durgan499 │ F │ Springfield │ X49016634X │
+│ Ty725 │ Schmeler639 │ M │ Springfield │ X22960735X │
+│ Pilar644 │ Pouros728 │ F │ Springfield │ X21434326X │
+│ Georgette866 │ Stark857 │ F │ Springfield │ X84034866X │
+│ Annika454 │ Gutmann970 │ F │ Springfield │ X3275916X │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Isabel214 │ Camacho176 │ F │ Springfield │ X52173808X │
+│ Carla633 │ Terán294 │ F │ Springfield │ X50212869X │
+│ Marceline716 │ Kuhlman484 │ F │ Springfield │ X70432308X │
+│ Ana María762 │ Cotto891 │ F │ Springfield │ X4604857X │
+│ Josefina523 │ Vanegas191 │ F │ Springfield │ X85396222X │
+│ Ester635 │ Sevilla788 │ F │ Springfield │ X50601786X │
+│ Tomás404 │ Galarza986 │ M │ Springfield │ X76790663X │
+│ Arturo47 │ Delafuente833 │ M │ Springfield │ X45087804X │
+│ Gilberto712 │ Martínez540 │ M │ Springfield │ X89729075X │
+│ Juanita470 │ Connelly992 │ F │ Springfield │ X71156217X │
+├──────────────┴────────────────┴─────────┴─────────────┴────────────┤
+│ 2221 rows (20 shown) 5 columns │
+└────────────────────────────────────────────────────────────────────┘
+```
+### Filter and select in numeric range
+Patients in City = 'Springfield' where the HEALTHCARE_EXPENSES between 1.5M and 2M
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients WHERE city = 'Springfield' AND HEALTHCARE_EXPENSES BETWEEN 1500000 AND 2000000;")
+```
+```python
+┌──────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────────┼────────────────┼─────────┼─────────────┤
+│ Talisha682 │ Brakus656 │ F │ Springfield │
+│ Jerold208 │ Harber290 │ M │ Springfield │
+│ Dean966 │ Tillman293 │ M │ Springfield │
+│ Orval846 │ Cartwright189 │ M │ Springfield │
+│ Jacinto644 │ Abernathy524 │ M │ Springfield │
+│ Bethel526 │ Satterfield305 │ F │ Springfield │
+│ Dorene845 │ Botsford977 │ F │ Springfield │
+│ Lula998 │ Langosh790 │ F │ Springfield │
+│ Deeanna316 │ Koss676 │ F │ Springfield │
+│ Loriann967 │ Torp761 │ F │ Springfield │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Ashley34 │ Murazik203 │ M │ Springfield │
+│ Ulysses632 │ Donnelly343 │ M │ Springfield │
+│ Horace32 │ Hammes673 │ M │ Springfield │
+│ Roman389 │ Lubowitz58 │ M │ Springfield │
+│ Jerald662 │ Grady603 │ M │ Springfield │
+│ Damien170 │ Hoppe518 │ M │ Springfield │
+│ Charley358 │ Vandervort697 │ M │ Springfield │
+│ Carla633 │ Terán294 │ F │ Springfield │
+│ Ana María762 │ Cotto891 │ F │ Springfield │
+│ Juanita470 │ Connelly992 │ F │ Springfield │
+├──────────────┴────────────────┴─────────┴─────────────┤
+│ 241 rows (20 shown) 4 columns │
+└───────────────────────────────────────────────────────┘
+```
+### LIMIT
+- LIMIT specifies the maximum number of records the query will return.
+
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients LIMIT 20;")
+```
+```python
+
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients LIMIT 20;")
+┌─────────────────┬────────────────┬─────────┬─────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├─────────────────┼────────────────┼─────────┼─────────────┤
+│ Jacinto644 │ Kris249 │ M │ Springfield │
+│ Alva958 │ Krajcik437 │ F │ Walpole │
+│ Jimmie93 │ Harris789 │ F │ Pembroke │
+│ Gregorio366 │ Auer97 │ M │ Boston │
+│ Karyn217 │ Mueller846 │ F │ Colrain │
+│ Jayson808 │ Fadel536 │ M │ Chicopee │
+│ José Eduardo181 │ Gómez206 │ M │ Chicopee │
+│ Milo271 │ Feil794 │ M │ Somerville │
+│ Karyn217 │ Metz686 │ F │ Medfield │
+│ Jeffrey461 │ Greenfelder433 │ M │ Springfield │
+│ Mariana775 │ Gulgowski816 │ F │ Lowell │
+│ Leann224 │ Deckow585 │ F │ Needham │
+│ Isabel214 │ Hinojosa147 │ F │ Fall River │
+│ Christal240 │ Brown30 │ F │ Boston │
+│ Carmelia328 │ Konopelski743 │ F │ Ashland │
+│ Raye931 │ Wyman904 │ F │ Quincy │
+│ Lisbeth69 │ Rowe323 │ F │ Malden │
+│ Amada498 │ Spinka232 │ F │ Foxborough │
+│ Cythia210 │ Reichel38 │ F │ Peabody │
+│ María Soledad68 │ Aparicio848 │ F │ Boston │
+├─────────────────┴────────────────┴─────────┴─────────────┤
+│ 20 rows 4 columns │
+└──────────────────────────────────────────────────────────┘
+```
+### ORDER BY and ASC
+This query returns the first 20 patients from the patients table, ordered alphabetically by their last name.
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients ORDER BY LAST ASC LIMIT 20;")
+```
+```python
+┌────────────────┬───────────┬─────────┬──────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├────────────────┼───────────┼─────────┼──────────────┤
+│ Jc393 │ Abbott774 │ M │ Westfield │
+│ Lloyd546 │ Abbott774 │ M │ Northampton │
+│ Charles364 │ Abbott774 │ M │ Brockton │
+│ Lelia627 │ Abbott774 │ F │ Gloucester │
+│ Devorah937 │ Abbott774 │ F │ Cambridge │
+│ Warren653 │ Abbott774 │ M │ Tyngsborough │
+│ Rhona164 │ Abbott774 │ F │ Stoneham │
+│ Jackqueline794 │ Abbott774 │ F │ Randolph │
+│ Willa615 │ Abbott774 │ F │ Dartmouth │
+│ Cyril535 │ Abbott774 │ M │ Millis │
+│ Lorette239 │ Abbott774 │ F │ Dennis │
+│ Jimmy858 │ Abbott774 │ M │ Lowell │
+│ Laine739 │ Abbott774 │ F │ Agawam │
+│ Bernetta267 │ Abbott774 │ F │ Ware │
+│ Lauri399 │ Abbott774 │ F │ Springfield │
+│ Miesha237 │ Abbott774 │ F │ Stoneham │
+│ Darrin898 │ Abbott774 │ M │ Newton │
+│ Grant908 │ Abbott774 │ M │ Arlington │
+│ Arden380 │ Abbott774 │ M │ Worcester │
+│ German382 │ Abbott774 │ M │ Taunton │
+├────────────────┴───────────┴─────────┴──────────────┤
+│ 20 rows 4 columns │
+└─────────────────────────────────────────────────────┘
+```
+### ORDER BY and DESC
+This query returns the first 20 patients from the patients table, ordered reverse alphabetically by their first name.
+```python
+conn.sql("SELECT FIRST, LAST, GENDER, CITY FROM patients ORDER BY FIRST DESC LIMIT 20;")
+```
+```python
+┌──────────┬────────────────┬─────────┬────────────┐
+│ FIRST │ LAST │ GENDER │ CITY │
+│ varchar │ varchar │ varchar │ varchar │
+├──────────┼────────────────┼─────────┼────────────┤
+│ Óscar156 │ Ballesteros368 │ M │ Taunton │
+│ Óscar156 │ Curiel392 │ M │ Swampscott │
+│ Óscar156 │ Zelaya592 │ M │ Lynn │
+│ Óscar156 │ Puente961 │ M │ Greenfield │
+│ Óscar156 │ Olivas524 │ M │ Norwell │
+│ Óscar156 │ Rivero165 │ M │ Lancaster │
+│ Óscar156 │ Canales95 │ M │ Hamilton │
+│ Óscar156 │ Romero158 │ M │ Boston │
+│ Óscar156 │ Garza151 │ M │ Longmeadow │
+│ Óscar156 │ Delgado712 │ M │ Haverhill │
+│ Óscar156 │ Ureña88 │ M │ Bedford │
+│ Óscar156 │ Henríquez109 │ M │ Boxborough │
+│ Óscar156 │ Guerrero997 │ M │ Boston │
+│ Óscar156 │ Ojeda263 │ M │ Boston │
+│ Óscar156 │ Meléndez48 │ M │ Newton │
+│ Óscar156 │ Muro989 │ M │ Boston │
+│ Óscar156 │ Rendón540 │ M │ Wilbraham │
+│ Óscar156 │ Santacruz647 │ M │ Winthrop │
+│ Óscar156 │ Santacruz647 │ M │ Boston │
+│ Óscar156 │ Otero621 │ M │ Boston │
+├──────────┴────────────────┴─────────┴────────────┤
+│ 20 rows 4 columns │
+└──────────────────────────────────────────────────┘
+```
+## Aggregating Data using GROUP BY
+#### Counting Patients by City
+
+```python
+conn.sql("SELECT CITY, COUNT(*) AS patient_count FROM patients GROUP BY CITY;") ## each run gives random order
+```
+```python
+┌─────────────┬───────────────┐
+│ CITY │ patient_count │
+│ varchar │ int64 │
+├─────────────┼───────────────┤
+│ Dartmouth │ 656 │
+│ Fitchburg │ 743 │
+│ Plymouth │ 972 │
+│ Worcester │ 3263 │
+│ Beverly │ 778 │
+│ Westfield │ 781 │
+│ Hingham │ 431 │
+│ Rochester │ 103 │
+│ Sandwich │ 387 │
+│ Watertown │ 534 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Ashfield │ 29 │
+│ Pelham │ 26 │
+│ Hancock │ 22 │
+│ Monterey │ 13 │
+│ Richmond │ 23 │
+│ Hinsdale │ 30 │
+│ Middlefield │ 11 │
+│ Westhampton │ 14 │
+│ Sandisfield │ 12 │
+│ Monroe │ 2 │
+├─────────────┴───────────────┤
+│ 351 rows (20 shown) │
+└─────────────────────────────┘
+```
+### Counting Patients by City and sorting them highest to lowest
+```python
+conn.sql("SELECT CITY, COUNT(*) AS patient_count FROM patients GROUP BY CITY ORDER BY patient_count DESC;")
+```
+```python
+┌─────────────┬───────────────┐
+│ CITY │ patient_count │
+│ varchar │ int64 │
+├─────────────┼───────────────┤
+│ Boston │ 11496 │
+│ Worcester │ 3263 │
+│ Springfield │ 2814 │
+│ Cambridge │ 2044 │
+│ Lowell │ 2027 │
+│ Brockton │ 1833 │
+│ Lynn │ 1714 │
+│ New Bedford │ 1706 │
+│ Quincy │ 1698 │
+│ Newton │ 1695 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Blandford │ 10 │
+│ Heath │ 8 │
+│ Tolland │ 7 │
+│ Hawley │ 6 │
+│ Alford │ 5 │
+│ New Ashford │ 4 │
+│ Aquinnah │ 4 │
+│ Tyringham │ 3 │
+│ Monroe │ 2 │
+│ Gosnold │ 1 │
+├─────────────┴───────────────┤
+│ 351 rows (20 shown) │
+└─────────────────────────────┘
+```
+### Total Number of Patients by Gender in Each City
+```python
+conn.sql("SELECT CITY, GENDER, COUNT(*) AS total_patients FROM patients GROUP BY CITY, GENDER ORDER BY total_patients DESC LIMIT 10;")
+```
+```python
+┌─────────────┬─────────┬────────────────┐
+│ CITY │ GENDER │ total_patients │
+│ varchar │ varchar │ int64 │
+├─────────────┼─────────┼────────────────┤
+│ Boston │ F │ 5840 │
+│ Boston │ M │ 5656 │
+│ Worcester │ M │ 1694 │
+│ Worcester │ F │ 1569 │
+│ Springfield │ M │ 1463 │
+│ Springfield │ F │ 1351 │
+│ Cambridge │ M │ 1023 │
+│ Cambridge │ F │ 1021 │
+│ Lowell │ F │ 1015 │
+│ Lowell │ M │ 1012 │
+├─────────────┴─────────┴────────────────┤
+│ 10 rows 3 columns │
+└────────────────────────────────────────┘
+```
+Lets look at other Tables - observations and immunizations
+```python
+conn.sql("SELECT PATIENT,ENCOUNTER,CODE, DESCRIPTION, VALUE from observations;")
+```
+```python
+┌──────────────────────┬──────────────────────┬─────────┬───────────────────────────────────────────────┬──────────────┐
+│ PATIENT │ ENCOUNTER │ CODE │ DESCRIPTION │ VALUE │
+│ varchar │ varchar │ varchar │ varchar │ varchar │
+├──────────────────────┼──────────────────────┼─────────┼───────────────────────────────────────────────┼──────────────┤
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8302-2 │ Body Height │ 82.7 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 72514-3 │ Pain severity - 0-10 verbal numeric rating … │ 2.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 29463-7 │ Body Weight │ 11.5 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 77606-2 │ Weight-for-length Per age and sex │ 47.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 9843-4 │ Head Occipital-frontal circumference │ 46.9 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8462-4 │ Diastolic Blood Pressure │ 76.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8480-6 │ Systolic Blood Pressure │ 107.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 8867-4 │ Heart rate │ 68.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 9279-1 │ Respiratory rate │ 13.0 │
+│ 1ff7f10f-a204-4bb1… │ 52051c30-c6c3-45fe… │ 72166-2 │ Tobacco smoking status NHIS │ Never smoker │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2160-0 │ Creatinine [Mass/volume] in Serum or Plasma │ 2.9 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 17861-6 │ Calcium [Mass/volume] in Serum or Plasma │ 9.2 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2951-2 │ Sodium [Moles/volume] in Serum or Plasma │ 143.8 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2823-3 │ Potassium [Moles/volume] in Serum or Plasma │ 4.3 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2075-0 │ Chloride [Moles/volume] in Serum or Plasma │ 106.4 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2028-9 │ Carbon dioxide total [Moles/volume] in Ser… │ 22.9 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 33914-3 │ Glomerular filtration rate/1.73 sq M.predic… │ 9.9 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 2885-2 │ Protein [Mass/volume] in Serum or Plasma │ 5.7 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 1751-7 │ Albumin [Mass/volume] in Serum or Plasma │ 5.2 │
+│ 87537cb1-92e1-4a11… │ 69e8eaf0-7714-4c20… │ 1975-2 │ Bilirubin.total [Mass/volume] in Serum or P… │ 14.2 │
+├──────────────────────┴──────────────────────┴─────────┴───────────────────────────────────────────────┴──────────────┤
+│ ? rows (>9999 rows, 20 shown) 5 columns │
+└──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+#### Average Heart Rate by Patient
+
+```python
+conn.sql("SELECT PATIENT, ROUND(AVG(CAST(VALUE AS DECIMAL(10,6))), 2) AS average_heart_rate FROM observations WHERE DESCRIPTION LIKE '%Heart rate%' GROUP BY PATIENT ORDER BY average_heart_rate DESC LIMIT 20;")
+```
+```python
+┌──────────────────────────────────────┬────────────────────┐
+│ PATIENT │ average_heart_rate │
+│ varchar │ double │
+├──────────────────────────────────────┼────────────────────┤
+│ ff2209f2-a4c9-4737-9aa7-adc8d71b6961 │ 200.0 │
+│ 8cea2d7e-6227-4924-a340-18eafb8564ac │ 200.0 │
+│ 6ec823e7-839e-4491-8b6f-6b275951b456 │ 200.0 │
+│ 0191f8aa-96dc-41e6-b718-312686a7a867 │ 200.0 │
+│ c10db4e1-de51-495f-98ab-8322c6d9550a │ 200.0 │
+│ 46ea7226-63f6-404f-bbed-904ea39f706d │ 199.9 │
+│ ec1ce32c-57a3-44c4-8f64-e050f2f94e03 │ 199.9 │
+│ a05e3bd1-1c51-453a-9969-646cb0168d23 │ 199.9 │
+│ 9cc52681-4446-4e4a-80be-63418dd06e66 │ 199.9 │
+│ b0112890-a24a-4564-b2b3-9b77879e5e79 │ 199.9 │
+│ 18f53179-2449-47af-b4da-a9e51095076f │ 199.9 │
+│ c3d165d2-597e-45a8-8331-f3338ce19fdf │ 199.9 │
+│ 04dd3439-2116-4f9a-b40e-a84395cf6f21 │ 199.9 │
+│ 371e14a1-31bd-4407-bdbb-4078781fde05 │ 199.8 │
+│ 3e5c63bf-65fe-48bb-8361-6b21a909888a │ 199.8 │
+│ 423f79cf-f7b8-41cd-bb3c-8459084f88eb │ 199.8 │
+│ 2d4f57ee-4001-44f8-9a94-c1885f26b408 │ 199.8 │
+│ bd6784c0-3529-4a12-87bd-746affb17739 │ 199.8 │
+│ ef43848b-a4f1-4c45-a106-5928e29b72f2 │ 199.7 │
+│ 1d050245-097c-44a5-b29b-6a18a22731e9 │ 199.7 │
+├──────────────────────────────────────┴────────────────────┤
+│ 20 rows 2 columns │
+└───────────────────────────────────────────────────────────┘
+```
+#### Count of Heart Rate Records by Patient
+```python
+conn.sql("SELECT PATIENT, COUNT(*) AS heart_rate_records FROM observations WHERE DESCRIPTION LIKE '%Heart rate%' GROUP BY PATIENT ORDER BY heart_rate_records DESC LIMIT 20;")
+```
+```python
+┌──────────────────────────────────────┬────────────────────┐
+│ PATIENT │ heart_rate_records │
+│ varchar │ int64 │
+├──────────────────────────────────────┼────────────────────┤
+│ 3e21a156-da54-4fb7-815e-550fdf4afbbd │ 33 │
+│ ffbf0392-1643-4b05-819f-489072c8c4d4 │ 30 │
+│ b9ef6005-438e-4b47-afaf-9dba32184adf │ 30 │
+│ f8080701-2b5c-4128-9a7f-9a098b737b27 │ 30 │
+│ a2172279-3d63-4d14-867d-1a39d9280690 │ 30 │
+│ 01a5a5a6-ef7b-42ba-899c-de66e1b1e27e │ 30 │
+│ 62572c44-a802-40d1-8d60-d02bc287d548 │ 30 │
+│ 445953fd-15fa-424e-9926-f93ebf1bca7a │ 30 │
+│ c3ea3c46-f8d1-4abe-8fb2-8fc8f86b4ef4 │ 29 │
+│ bd18ba0d-2e65-4427-98f1-cbfc09dbaa33 │ 29 │
+│ 5736b489-0e15-4693-b9d6-05192a702a2d │ 29 │
+│ b5a0b060-96ae-44d5-bd10-f532a445009b │ 29 │
+│ ee9b44a1-36c6-456e-b77c-cd200611ca0f │ 29 │
+│ 0d5290dd-a3e3-4a81-91d5-6e722e58ec3e │ 29 │
+│ 54ec8b8e-fe2a-4cf5-a087-1a65df1ab1b8 │ 29 │
+│ e658626d-0f72-4570-8071-2dd63827a24c │ 29 │
+│ ec3f3da2-09a1-43c7-8cae-ed34847d2534 │ 28 │
+│ c7bd54b1-e036-4702-9062-acfa3677a6e8 │ 28 │
+│ 8b6ee161-eb5f-413a-8e86-f5b3adb3f5ff │ 28 │
+│ 03c1210e-d145-4aed-9522-14cff41b3c28 │ 28 │
+├──────────────────────────────────────┴────────────────────┤
+│ 20 rows 2 columns │
+└───────────────────────────────────────────────────────────┘
+```
+#### Total Cost of Vaccinations per Patient
+```python
+conn.sql("SELECT PATIENT, SUM(BASE_COST) AS total_cost FROM immunizations GROUP BY PATIENT ORDER BY total_cost DESC;")
+```
+```python
+┌──────────────────────────────────────┬────────────┐
+│ PATIENT │ total_cost │
+│ varchar │ double │
+├──────────────────────────────────────┼────────────┤
+│ 8eacdc17-de2f-4024-88bd-976401b30979 │ 2810.4 │
+│ bb14de6a-c77d-44bb-a3a8-338a6a246520 │ 2810.4 │
+│ a5b60d43-9776-4811-badf-67d49edbc175 │ 2810.4 │
+│ 1c8a4026-5dbe-478a-b647-ecb605775977 │ 2810.4 │
+│ 3e38e20b-6dd8-44fe-9ed3-81e1e3d8e2ab │ 2810.4 │
+│ e23c7c68-0319-41f5-90d9-1db160321a94 │ 2810.4 │
+│ 49e7d878-93a6-4e71-971b-213abacf2235 │ 2810.4 │
+│ 7ded4116-1783-4a93-9bb2-7a65370f55f5 │ 2810.4 │
+│ 594660e4-6a02-4e3c-a8b9-2c61d4d4af60 │ 2810.4 │
+│ fee1edbd-277d-4b12-901e-60b6135cf877 │ 2810.4 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ 594d1ecb-b1a4-4bad-9ccd-5cebceebd6f5 │ 281.04 │
+│ 456fa9f9-7eb7-4316-a74b-5a9ff87f8fc5 │ 281.04 │
+│ 52568520-7163-429d-bc90-fc245debb697 │ 281.04 │
+│ 30a16a19-ee93-4bfd-b1ed-bba90de36266 │ 281.04 │
+│ 0ef633b0-2996-44c5-936c-600820310223 │ 281.04 │
+│ 613319c8-2b76-494b-86b8-e776a2ee5e0b │ 281.04 │
+│ 4dceeb57-4fe3-4c32-a748-621df5ce3c30 │ 281.04 │
+│ 93d590e8-9f3e-4741-bee2-9b8e9e8b21e7 │ 281.04 │
+│ bad20ffc-48f9-4a7d-a2be-39a50fa4e0ac │ 281.04 │
+│ 3cf75ec3-5e1c-4783-bb20-282a8cf66f30 │ 281.04 │
+├──────────────────────────────────────┴────────────┤
+│ ? rows (>9999 rows, 20 shown) 2 columns │
+└───────────────────────────────────────────────────┘
+```
+#### Top 10 Most Common Vaccine Administered
+```python
+conn.sql("SELECT DESCRIPTION, COUNT(*) AS count FROM immunizations GROUP BY DESCRIPTION ORDER BY count DESC LIMIT 10;")
+```
+```python
+┌────────────────────────────────────────────────────┬────────┐
+│ DESCRIPTION │ count │
+│ varchar │ int64 │
+├────────────────────────────────────────────────────┼────────┤
+│ Influenza seasonal injectable preservative free │ 106564 │
+│ Td (adult) preservative free │ 9815 │
+│ Pneumococcal conjugate PCV 13 │ 5747 │
+│ DTaP │ 5735 │
+│ IPV │ 4962 │
+│ meningococcal MCV4P │ 4010 │
+│ Hib (PRP-OMP) │ 3615 │
+│ HPV quadrivalent │ 3494 │
+│ Hep B adolescent or pediatric │ 3490 │
+│ zoster │ 3469 │
+├────────────────────────────────────────────────────┴────────┤
+│ 10 rows 2 columns │
+└─────────────────────────────────────────────────────────────┘
+```
+## JOINS
+#### SQL joins are used to combine rows from two or more tables, based on a related column between them. There are several types of joins in SQL, each serving different purposes depending on how you want to combine your data.
+
+
+ - credit: educba.com
+
+___
+
+For the following examples: Left table: Patients and Right Table: Immunizations
+
+__
+
+### INNER JOIN
+- The Inner join returns rows when there is at least one match in both tables. If there is no match, the rows are not returned.
+
+- Find Matching Records: Question: Which medical treatments have been administered to patients, including the patient's name and the cost of each treatment?
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM immunizations INNER JOIN patients ON immunizations.PATIENT = patients.Id ORDER BY patients.FIRST ASC;")
+```
+```python
+┌──────────┬───────────────┬────────────────────────────────────────────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├──────────┼───────────────┼────────────────────────────────────────────────────┼───────────┤
+│ Aaron697 │ Cummings51 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Anderson154 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Aaron697 │ Hartmann983 │ Hep B adolescent or pediatric │ 140.52 │
+│ Aaron697 │ Thompson596 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Rodriguez71 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Deckow585 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Swaniawski813 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Beer512 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Ullrich385 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Ariel183 │ Schuster709 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Mohr916 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Waters156 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Will178 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Leffler128 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Bode78 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Grady603 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ Funk324 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Ariel183 │ McGlynn426 │ Td (adult) preservative free │ 140.52 │
+│ Ariel183 │ Boehm581 │ Influenza seasonal injectable preservative free │ 140.52 │
+├──────────┴───────────────┴────────────────────────────────────────────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└───────────────────────────────────────────────────────────────────────────────────────────┘
+```
+
+### LEFT JOIN
+- It returns all rows from the left table, and the matched rows from the right table. The result is NULL from the right side, if there is no match.
+Question: For all patients, what treatments have they received, if any?
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients LEFT JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY patients.FIRST ASC;")
+```
+```python
+
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients LEFT JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY patients.FIRST ASC;")
+┌───────────┬─────────────┬────────────────────────────────────────────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├───────────┼─────────────┼────────────────────────────────────────────────────┼───────────┤
+│ Aaron697 │ Legros616 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Deckow585 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Thompson596 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Auer97 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Td (adult) preservative free │ 140.52 │
+│ Aaron697 │ Volkman526 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Hartmann983 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Orn563 │ NULL │ NULL │
+│ Aaron697 │ Hartmann983 │ IPV │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Antonia30 │ Goodwin327 │ HPV quadrivalent │ 140.52 │
+│ Antonia30 │ Funk324 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ West559 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Lowe577 │ NULL │ NULL │
+│ Antonia30 │ D'Amore443 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Montes106 │ meningococcal MCV4P │ 140.52 │
+│ Antonia30 │ González124 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Franecki195 │ zoster │ 140.52 │
+│ Antonia30 │ Rojo930 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Puente961 │ HPV quadrivalent │ 140.52 │
+├───────────┴─────────────┴────────────────────────────────────────────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────────────────────────────────────┘
+```
+### RIGHT JOIN
+- It returns all rows from the right table, and the matched rows from the left table. The result is NULL from the left side, if there is no match.
+Question: For all Immunizations recorded, which patients received them, if identifiable?
+
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients RIGHT JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY immunizations.DESCRIPTION;")
+```
+```python
+┌────────────────┬──────────────┬──────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├────────────────┼──────────────┼──────────────┼───────────┤
+│ Shantay950 │ Quitzon246 │ DTaP │ 140.52 │
+│ Cleveland582 │ VonRueden376 │ DTaP │ 140.52 │
+│ Flo729 │ Quigley282 │ DTaP │ 140.52 │
+│ Flo729 │ Quigley282 │ DTaP │ 140.52 │
+│ Werner409 │ Schaden604 │ DTaP │ 140.52 │
+│ Pasty639 │ Ortiz186 │ DTaP │ 140.52 │
+│ Pasty639 │ Ortiz186 │ DTaP │ 140.52 │
+│ Angelica442 │ Kovacek682 │ DTaP │ 140.52 │
+│ Cathie710 │ Hegmann834 │ DTaP │ 140.52 │
+│ Pasty639 │ Ortiz186 │ DTaP │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Art115 │ Roberts511 │ Hep A adult │ 140.52 │
+│ Christopher407 │ Davis923 │ Hep A adult │ 140.52 │
+│ Michiko564 │ Dooley940 │ Hep A adult │ 140.52 │
+│ Celine582 │ Sipes176 │ Hep A adult │ 140.52 │
+│ Darci883 │ Miller503 │ Hep A adult │ 140.52 │
+│ Ethan766 │ Morar593 │ Hep A adult │ 140.52 │
+│ Georgiann138 │ Heathcote539 │ Hep A adult │ 140.52 │
+│ Herbert830 │ Wolff180 │ Hep A adult │ 140.52 │
+│ Ismael683 │ King743 │ Hep A adult │ 140.52 │
+│ Ellsworth48 │ Mertz280 │ Hep A adult │ 140.52 │
+├────────────────┴──────────────┴──────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────┘
+```
+### FULL OUTER JOIN
+- It returns rows when there is a match in one of the tables. It effectively combines the results of both LEFT JOIN and RIGHT JOIN.
+Question: What is the complete list of patients and their Immunizations, including those without recorded treatments or identifiable patients?
+
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION, immunizations.BASE_COST FROM patients FULL OUTER JOIN immunizations ON patients.Id = immunizations.PATIENT ORDER BY patients.FIRST ASC;")
+```
+```python
+┌───────────┬─────────────┬────────────────────────────────────────────────────┬───────────┐
+│ FIRST │ LAST │ DESCRIPTION │ BASE_COST │
+│ varchar │ varchar │ varchar │ double │
+├───────────┼─────────────┼────────────────────────────────────────────────────┼───────────┤
+│ Aaron697 │ Legros616 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Deckow585 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Thompson596 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Auer97 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Crooks415 │ Td (adult) preservative free │ 140.52 │
+│ Aaron697 │ Volkman526 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Hartmann983 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Aaron697 │ Orn563 │ NULL │ NULL │
+│ Aaron697 │ Hartmann983 │ IPV │ 140.52 │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Antonia30 │ Goodwin327 │ HPV quadrivalent │ 140.52 │
+│ Antonia30 │ Funk324 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ West559 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Lowe577 │ NULL │ NULL │
+│ Antonia30 │ D'Amore443 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Montes106 │ meningococcal MCV4P │ 140.52 │
+│ Antonia30 │ González124 │ Influenza seasonal injectable preservative free │ 140.52 │
+│ Antonia30 │ Franecki195 │ zoster │ 140.52 │
+│ Antonia30 │ Rojo930 │ Pneumococcal conjugate PCV 13 │ 140.52 │
+│ Antonia30 │ Puente961 │ HPV quadrivalent │ 140.52 │
+├───────────┴─────────────┴────────────────────────────────────────────────────┴───────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└──────────────────────────────────────────────────────────────────────────────────────────┘
+```
+### CROSS JOIN
+- It returns a Cartesian product of the two tables, i.e., it returns rows combining each row from the first table with each row from the second table.
+Question: What are all possible combinations of patients and immunizations?
+```python
+conn.sql("SELECT patients.FIRST, patients.LAST, immunizations.DESCRIPTION FROM patients CROSS JOIN immunizations;")
+```
+```python
+┌───────────┬─────────────┬────────────────────────────────────────────────────┐
+│ FIRST │ LAST │ DESCRIPTION │
+│ varchar │ varchar │ varchar │
+├───────────┼─────────────┼────────────────────────────────────────────────────┤
+│ Tammy740 │ Ernser583 │ Influenza seasonal injectable preservative free │
+│ Tammy740 │ Ernser583 │ Hep A ped/adol 2 dose │
+│ Tammy740 │ Ernser583 │ Influenza seasonal injectable preservative free │
+│ Tammy740 │ Ernser583 │ Influenza seasonal injectable preservative free │
+│ Tammy740 │ Ernser583 │ meningococcal MCV4P │
+│ Tammy740 │ Ernser583 │ Hep B adolescent or pediatric │
+│ Tammy740 │ Ernser583 │ Hep B adolescent or pediatric │
+│ Tammy740 │ Ernser583 │ Hib (PRP-OMP) │
+│ Tammy740 │ Ernser583 │ rotavirus monovalent │
+│ Tammy740 │ Ernser583 │ IPV │
+│ · │ · │ · │
+│ · │ · │ · │
+│ · │ · │ · │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ zoster │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+│ Iliana226 │ Schmeler639 │ Influenza seasonal injectable preservative free │
+├───────────┴─────────────┴────────────────────────────────────────────────────┤
+│ ? rows (>9999 rows, 20 shown) 3 columns │
+└──────────────────────────────────────────────────────────────────────────────┘
+```
+### SELF JOIN
+- It is not a different type of join, but it's a regular join used to join a table to itself. It's useful for queries where you need to compare rows within the same table.
+Question: Which pair of patients are from the same city?
+```python
+conn.sql("SELECT A.FIRST AS FirstPatient, B.FIRST AS SecondPatient, A.CITY FROM patients A, patients B WHERE A.CITY = B.CITY AND A.Id != B.Id ORDER BY A.CITY ASC;")
+```
+```python
+┌──────────────┬───────────────┬──────────┐
+│ FirstPatient │ SecondPatient │ CITY │
+│ varchar │ varchar │ varchar │
+├──────────────┼───────────────┼──────────┤
+│ Susan422 │ Stefani254 │ Abington │
+│ Jennell254 │ Lionel365 │ Abington │
+│ Robbie31 │ Antonio44 │ Abington │
+│ Joe656 │ Antonio44 │ Abington │
+│ Susan422 │ Stevie682 │ Abington │
+│ Shaun461 │ Stevie682 │ Abington │
+│ Shaun461 │ Weston546 │ Abington │
+│ Susan422 │ Elwood28 │ Abington │
+│ Susan422 │ Daria61 │ Abington │
+│ Shaun461 │ Lita714 │ Abington │
+│ · │ · │ · │
+│ · │ · │ · │
+│ · │ · │ · │
+│ Elane105 │ Conrad619 │ Abington │
+│ Logan497 │ Conrad619 │ Abington │
+│ Sherlene302 │ Conrad619 │ Abington │
+│ Milan77 │ Conrad619 │ Abington │
+│ Frankie174 │ Seema671 │ Abington │
+│ Elane105 │ Josefina523 │ Abington │
+│ Lisha487 │ Shawanna357 │ Abington │
+│ Marleen824 │ Marcelino726 │ Abington │
+│ Sandi885 │ Lionel365 │ Abington │
+│ Morton637 │ Viva686 │ Abington │
+├──────────────┴───────────────┴──────────┤
+│ ? rows (>9999 rows, 20 shown) │
+└─────────────────────────────────────────┘
+```
+___
+___
+
+## Introduction to Subqueries and Nested Selects
+- Subqueries, also known as inner queries or nested queries, are SQL queries nested inside a larger query. They allow you to perform operations that usually require multiple steps in a single query, making your data retrieval process more efficient and concise. Nested selects are a type of subquery used specifically within the SELECT, FROM, or WHERE clauses to provide a dataset for the outer query to process.
+- Subqueries can return a single value, a single row, multiple rows, or a table. They are used for comparison, as a condition, or to provide a list of values for the outer query. The main distinction between correlated and non-correlated subqueries is that correlated subqueries reference column(s) from the outer query, thus running once for each row selected by the outer query, while non-correlated subqueries run independently of the outer query and can be run as standalone queries.
+
+### Types of subqueries
+* Single-Row Subqueries: Single-row subqueries return only one row and are used with single row comparison operators like =, >, <, >=, <=. They are often used in the WHERE clause to compare a column value against the result of the subquery.
+* Multi-Row Subqueries: Multi-row subqueries return more than one row and are used with operators like IN, ANY, ALL, which allow comparison against multiple values. They're useful for filtering based on a set of criteria returned by the subquery.
+* Correlated Subqueries: Correlated subqueries reference column(s) from the outer query, making them dependent on the outer query. They are executed once for each row processed by the outer query, often leading to performance considerations.
+
+___
+#### Use subqueries to find patients based on specific criteria.
+
+#### Find all patients who have been prescribed medication with a base cost higher than the average base cost of all medications.
+```python
+query = """SELECT p.FIRST, p.LAST
+FROM patients p
+WHERE p.Id IN ( SELECT m.PATIENT FROM medications m WHERE m.BASE_COST > (SELECT AVG(BASE_COST) FROM medications));
+"""
+conn.sql(query)
+```
+```python
+┌─────────────┬─────────────┐
+│ FIRST │ LAST │
+│ varchar │ varchar │
+├─────────────┼─────────────┤
+│ Dewitt635 │ Reichel38 │
+│ Anisa442 │ Purdy2 │
+│ Ross213 │ Mayert710 │
+│ Jim478 │ Mueller846 │
+│ Maria750 │ Schimmel440 │
+│ Ignacio928 │ Gorczany269 │
+│ Kip442 │ Zboncak558 │
+│ Jean712 │ Kuhlman484 │
+│ Mac103 │ Moen819 │
+│ Dianna917 │ Goldner995 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Cameron381 │ Bogan287 │
+│ Andrew29 │ Donnelly343 │
+│ Charis952 │ Littel644 │
+│ Lyndon118 │ Swift555 │
+│ Edward499 │ Zieme486 │
+│ Myron933 │ Ritchie586 │
+│ Yolonda722 │ Champlin946 │
+│ Alayna598 │ Kozey370 │
+│ Kristian973 │ Ledner144 │
+│ Sydney660 │ Zulauf375 │
+├─────────────┴─────────────┤
+│ ? rows 2 columns │
+└───────────────────────────┘
+```
+### Correlated Subqueries
+Learn how to use correlated subqueries to perform row-specific comparisons.
+Find patients whose healthcare expenses are higher than the average expenses in their county.
+```python
+query = """SELECT
+ p.FIRST,
+ p.LAST,
+ p.COUNTY,
+ p.HEALTHCARE_EXPENSES,
+ ROUND(AVG_EXPENSES.COUNTY_AVG_EXPENSES, 2) AS ROUNDED_AVG_HEALTHCARE_EXPENSES
+FROM
+ patients p
+INNER JOIN (
+ SELECT
+ COUNTY,
+ AVG(CAST(HEALTHCARE_EXPENSES AS DECIMAL(20,6))) AS COUNTY_AVG_EXPENSES
+ FROM
+ patients
+ GROUP BY
+ COUNTY
+) AS AVG_EXPENSES ON p.COUNTY = AVG_EXPENSES.COUNTY
+WHERE
+ p.HEALTHCARE_EXPENSES > (
+ SELECT AVG(p2.HEALTHCARE_EXPENSES)
+ FROM patients p2
+ WHERE p2.COUNTY = p.COUNTY
+ );
+"""
+conn.sql(query)
+```
+```python
+
+query = """SELECT
+ p.FIRST,
+ p.LAST,
+ p.COUNTY,
+ p.HEALTHCARE_EXPENSES,
+ ROUND(AVG_EXPENSES.COUNTY_AVG_EXPENSES, 2) AS ROUNDED_AVG_HEALTHCARE_EXPENSES
+FROM
+ patients p
+INNER JOIN (
+ SELECT
+… FROM patients p2
+ WHERE p2.COUNTY = p.COUNTY
+ );
+"""
+conn.sql(query)
+┌────────────┬───────────────┬───────────────────┬─────────────────────┬─────────────────────────────────┐
+│ FIRST │ LAST │ COUNTY │ HEALTHCARE_EXPENSES │ ROUNDED_AVG_HEALTHCARE_EXPENSES │
+│ varchar │ varchar │ varchar │ double │ double │
+├────────────┼───────────────┼───────────────────┼─────────────────────┼─────────────────────────────────┤
+│ Tammy740 │ Ernser583 │ Bristol County │ 1546025.67 │ 793296.73 │
+│ Iliana226 │ Schmeler639 │ Barnstable County │ 1407960.93 │ 949155.23 │
+│ Anthony633 │ Yundt842 │ Essex County │ 1575731.48 │ 814437.7 │
+│ Jim478 │ Mueller846 │ Bristol County │ 1112473.78 │ 793296.73 │
+│ Sina65 │ Howell947 │ Middlesex County │ 1479425.58 │ 841491.73 │
+│ Maria750 │ Schimmel440 │ Plymouth County │ 952814.55 │ 856778.1 │
+│ Lorenzo669 │ Vandervort697 │ Middlesex County │ 873001.47 │ 841491.73 │
+│ David908 │ Carter549 │ Worcester County │ 834576.02 │ 804752.74 │
+│ Noah480 │ O'Reilly797 │ Essex County │ 915110.6 │ 814437.7 │
+│ Kip442 │ Zboncak558 │ Middlesex County │ 1704103.7200000002 │ 841491.73 │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Tinisha932 │ Zulauf375 │ Bristol County │ 1035244.36 │ 793296.73 │
+│ Mario764 │ Estrada938 │ Suffolk County │ 1380401.5 │ 664340.79 │
+│ Glenn0 │ Schulist381 │ Barnstable County │ 1357123.8 │ 949155.23 │
+│ Porter490 │ Schulist381 │ Bristol County │ 1534216.57 │ 793296.73 │
+│ Darius626 │ Hackett68 │ Essex County │ 1715140.52 │ 814437.7 │
+│ Leonel449 │ Kunze215 │ Essex County │ 1492352.86 │ 814437.7 │
+│ Tyrell880 │ Kassulke119 │ Hampden County │ 1053520.21 │ 725636.71 │
+│ Arlen68 │ Gusikowski974 │ Worcester County │ 1363289.9 │ 804752.74 │
+│ Ashli227 │ Huel628 │ Worcester County │ 813730.02 │ 804752.74 │
+│ Loriann967 │ Mertz280 │ Norfolk County │ 1618397.57 │ 874351.83 │
+├────────────┴───────────────┴───────────────────┴─────────────────────┴─────────────────────────────────┤
+│ ? rows (>9999 rows, 20 shown) 5 columns │
+└────────────────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+Identify patients who have not been prescribed any medication.
+```python
+query = """
+SELECT p.FIRST, p.LAST
+FROM patients p
+WHERE NOT EXISTS (
+ SELECT 1 FROM medications m WHERE m.PATIENT = p.Id
+);
+"""
+
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT p.FIRST, p.LAST
+FROM patients p
+WHERE NOT EXISTS (
+ SELECT 1 FROM medications m WHERE m.PATIENT = p.Id
+);
+"""
+
+conn.sql(query)
+┌────────────┬───────────────┐
+│ FIRST │ LAST │
+│ varchar │ varchar │
+├────────────┼───────────────┤
+│ Iliana226 │ Schmeler639 │
+│ Toshiko149 │ Swaniawski813 │
+│ Heath320 │ Streich926 │
+│ Rana586 │ Langworth352 │
+│ Dorathy429 │ Yost751 │
+│ Deon400 │ Littel644 │
+│ Emerson869 │ D'Amore443 │
+│ Alec433 │ Sanford861 │
+│ Roselyn270 │ McLaughlin530 │
+│ Claudia969 │ Smith67 │
+│ · │ · │
+│ · │ · │
+│ · │ · │
+│ Lou594 │ Gleason633 │
+│ Hank686 │ Russel238 │
+│ Earl438 │ Block661 │
+│ Mariano761 │ Heller342 │
+│ Sandra485 │ Fisher429 │
+│ Claire652 │ Murray856 │
+│ Doreen575 │ Graham902 │
+│ Fausto876 │ Kuhn96 │
+│ Vernon254 │ Gutmann970 │
+│ Ernest565 │ Block661 │
+├────────────┴───────────────┤
+│ ? rows 2 columns │
+└────────────────────────────┘
+```
+Find patients whose healthcare coverage is above the average of their respective city.
+```python
+query = """
+SELECT
+ p.FIRST,
+ p.LAST,
+ p.CITY,
+ ROUND(p.HEALTHCARE_COVERAGE,2) AS INDIVIDUAL_COVERAGE,
+ ROUND(c.AVG_COVERAGE,2) AS CITY_AVERAGE_COVERAGE
+FROM
+ patients p
+INNER JOIN (
+ SELECT
+ CITY,
+ AVG(HEALTHCARE_COVERAGE) AS AVG_COVERAGE
+ FROM
+ patients
+ GROUP BY
+ CITY
+) AS c ON p.CITY = c.CITY
+WHERE
+ p.HEALTHCARE_COVERAGE > (
+ SELECT AVG(HEALTHCARE_COVERAGE)
+ FROM patients
+ WHERE CITY = p.CITY
+ );
+"""
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT
+ p.FIRST,
+ p.LAST,
+ p.CITY,
+ ROUND(p.HEALTHCARE_COVERAGE,2) AS INDIVIDUAL_COVERAGE,
+ ROUND(c.AVG_COVERAGE,2) AS CITY_AVERAGE_COVERAGE
+FROM
+ patients p
+INNER JOIN (
+… SELECT AVG(HEALTHCARE_COVERAGE)
+ FROM patients
+ WHERE CITY = p.CITY
+ );
+"""
+conn.sql(query)
+┌──────────────┬────────────────┬───────────────┬─────────────────────┬───────────────────────┐
+│ FIRST │ LAST │ CITY │ INDIVIDUAL_COVERAGE │ CITY_AVERAGE_COVERAGE │
+│ varchar │ varchar │ varchar │ double │ double │
+├──────────────┼────────────────┼───────────────┼─────────────────────┼───────────────────────┤
+│ Anisa442 │ Purdy2 │ Methuen │ 34393.57 │ 12949.31 │
+│ Sina65 │ Howell947 │ Sudbury │ 15481.43 │ 15385.85 │
+│ Maria750 │ Schimmel440 │ Plymouth │ 14231.22 │ 10826.53 │
+│ Kip442 │ Zboncak558 │ Hopkinton │ 155478.44 │ 15439.17 │
+│ Hans694 │ Wilkinson796 │ Framingham │ 13729.73 │ 11988.99 │
+│ Jean712 │ Kuhlman484 │ Boxford │ 11421.48 │ 8660.53 │
+│ Lorette239 │ Abbott774 │ Dennis │ 78740.23 │ 14918.28 │
+│ Tobi258 │ Bernier607 │ Wilmington │ 13889.6 │ 9411.52 │
+│ Rosia390 │ Reichel38 │ Rochester │ 16234.29 │ 9736.74 │
+│ Florencia449 │ Mercado213 │ Wilbraham │ 11427.25 │ 10468.26 │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ · │ · │ · │ · │ · │
+│ Jc393 │ Marks830 │ Brockton │ 16284.35 │ 12287.77 │
+│ Cedric746 │ Marvin195 │ Dedham │ 19806.72 │ 13840.99 │
+│ Milford485 │ Schmitt836 │ Hubbardston │ 10592.34 │ 8172.48 │
+│ Tegan755 │ Ruecker817 │ Waltham │ 30493.65 │ 14495.07 │
+│ Lindsay928 │ Lang846 │ Boston │ 11837.05 │ 10725.8 │
+│ Claudie965 │ Hermann103 │ Medford │ 15420.54 │ 12610.05 │
+│ Julie245 │ Nolan344 │ Essex │ 31903.53 │ 31787.13 │
+│ Mui729 │ Stoltenberg489 │ Wakefield │ 21528.91 │ 13699.32 │
+│ Oliver401 │ Lesch175 │ Yarmouth │ 27442.87 │ 19907.83 │
+│ Mi492 │ Gusikowski974 │ North Andover │ 18481.09 │ 17677.13 │
+├──────────────┴────────────────┴───────────────┴─────────────────────┴───────────────────────┤
+│ ? rows (>9999 rows, 20 shown) 5 columns │
+└─────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+## FUNCTIONS IN SQL
+___
+### String Functions
+### CONCAT: Concatenates two or more strings.
+```python
+query = """SELECT CONCAT(FIRST, ' ', LAST) AS FullName FROM patients LIMIT 10;
+;"""
+conn.sql(query)
+```
+```python
+┌───────────────────────────┐
+│ FullName │
+│ varchar │
+├───────────────────────────┤
+│ Jacinto644 Kris249 │
+│ Alva958 Krajcik437 │
+│ Jimmie93 Harris789 │
+│ Gregorio366 Auer97 │
+│ Karyn217 Mueller846 │
+│ Jayson808 Fadel536 │
+│ José Eduardo181 Gómez206 │
+│ Milo271 Feil794 │
+│ Karyn217 Metz686 │
+│ Jeffrey461 Greenfelder433 │
+├───────────────────────────┤
+│ 10 rows │
+└───────────────────────────┘
+```
+### DATALENGTH: Returns the number of bytes used to represent any expre
+```python
+query = """
+SELECT FIRST, LENGTH(FIRST) AS LengthInBytes FROM patients LIMIT 10;
+"""
+
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT FIRST, LENGTH(FIRST) AS LengthInBytes FROM patients LIMIT 10;
+"""
+
+conn.sql(query)
+┌─────────────────┬───────────────┐
+│ FIRST │ LengthInBytes │
+│ varchar │ int64 │
+├─────────────────┼───────────────┤
+│ Jacinto644 │ 10 │
+│ Alva958 │ 7 │
+│ Jimmie93 │ 8 │
+│ Gregorio366 │ 11 │
+│ Karyn217 │ 8 │
+│ Jayson808 │ 9 │
+│ José Eduardo181 │ 15 │
+│ Milo271 │ 7 │
+│ Karyn217 │ 8 │
+│ Jeffrey461 │ 10 │
+├─────────────────┴───────────────┤
+│ 10 rows 2 columns │
+└─────────────────────────────────┘
+```
+### LEFT: Returns the left part of a character string with the specified number of characters.
+```python
+query = """SELECT LEFT(FIRST, 5) AS Initial FROM patients LIMIT 10;"""
+conn.sql(query)
+```
+```python
+
+query = """SELECT LEFT(FIRST, 5) AS Initial FROM patients LIMIT 10;"""
+conn.sql(query)
+┌─────────┐
+│ Initial │
+│ varchar │
+├─────────┤
+│ Jacin │
+│ Alva9 │
+│ Jimmi │
+│ Grego │
+│ Karyn │
+│ Jayso │
+│ José │
+│ Milo2 │
+│ Karyn │
+│ Jeffr │
+├─────────┤
+│ 10 rows │
+└─────────┘
+```
+### LOWER: Converts all characters in the specified string to lowercase.
+### UPPER: Convert to uppercase
+```python
+query = """
+SELECT LOWER(FIRST) AS LowercaseFirstName, UPPER(LAST) AS UppercaseLastName FROM patients LIMIT 10;
+"""
+conn.sql(query)
+```
+```python
+
+query = """
+SELECT LOWER(FIRST) AS LowercaseFirstName, UPPER(LAST) AS UppercaseLastName FROM patients LIMIT 10;
+"""
+conn.sql(query)
+┌────────────────────┬───────────────────┐
+│ LowercaseFirstName │ UppercaseLastName │
+│ varchar │ varchar │
+├────────────────────┼───────────────────┤
+│ jacinto644 │ KRIS249 │
+│ alva958 │ KRAJCIK437 │
+│ jimmie93 │ HARRIS789 │
+│ gregorio366 │ AUER97 │
+│ karyn217 │ MUELLER846 │
+│ jayson808 │ FADEL536 │
+│ josé eduardo181 │ GÓMEZ206 │
+│ milo271 │ FEIL794 │
+│ karyn217 │ METZ686 │
+│ jeffrey461 │ GREENFELDER433 │
+├────────────────────┴───────────────────┤
+│ 10 rows 2 columns │
+└────────────────────────────────────────┘
+```
+LTRIM: Removes leading spaces from a string.
+```python
+query = """SELECT LTRIM(ADDRESS) AS TrimmedAddress FROM patients LIMIT 10;"""
+
+conn.sql(query)
+
+```
+```python
+
+query = """SELECT LTRIM(ADDRESS) AS TrimmedAddress FROM patients LIMIT 10;"""
+
+conn.sql(query)
+
+┌────────────────────────────────┐
+│ TrimmedAddress │
+│ varchar │
+├────────────────────────────────┤
+│ 888 Hickle Ferry Suite 38 │
+│ 1048 Skiles Trailer │
+│ 201 Mitchell Lodge Unit 67 │
+│ 1050 Lindgren Extension Apt 38 │
+│ 570 Abshire Forge Suite 32 │
+│ 1056 Harris Lane Suite 70 │
+│ 427 Balistreri Way Unit 19 │
+│ 422 Farrell Path Unit 69 │
+│ 181 Feest Passage Suite 64 │
+│ 428 Wiza Glen Unit 91 │
+├────────────────────────────────┤
+│ 10 rows │
+└────────────────────────────────┘
+```
+REPLACE: Replaces all occurrences of a specified string value with another string value.
+```python
+query = """SELECT FIRST, LAST,
+ ADDRESS AS LongAddress,
+ REPLACE(ADDRESS, 'Street', 'St') AS ShortAddress
+FROM
+ patients
+WHERE
+ ADDRESS LIKE '%Street%' LIMIT 10;
+"""
+
+conn.sql(query)
+```
+```python
+┌────────────┬───────────────┬───────────────────────────┬───────────────────────┐
+│ FIRST │ LAST │ LongAddress │ ShortAddress │
+│ varchar │ varchar │ varchar │ varchar │
+├────────────┼───────────────┼───────────────────────────┼───────────────────────┤
+│ Lorrie905 │ Leannon79 │ 813 Casper Street │ 813 Casper St │
+│ Asa127 │ Block661 │ 140 Rohan Street Suite 50 │ 140 Rohan St Suite 50 │
+│ Logan497 │ Brekke496 │ 1081 Orn Street │ 1081 Orn St │
+│ Cletus494 │ Strosin214 │ 1019 Haley Street │ 1019 Haley St │
+│ Cortney940 │ Stehr398 │ 458 Streich Street │ 458 Streich St │
+│ Latoyia537 │ Gaylord332 │ 200 Heaney Street │ 200 Heaney St │
+│ Clark193 │ Hilll811 │ 202 Tromp Street Suite 0 │ 202 Tromp St Suite 0 │
+│ Jc393 │ Bosco882 │ 792 Walsh Street │ 792 Walsh St │
+│ Noel608 │ Swaniawski813 │ 155 Walter Street │ 155 Walter St │
+│ Sari509 │ Hoppe518 │ 550 Lang Street Suite 65 │ 550 Lang St Suite 65 │
+├────────────┴───────────────┴───────────────────────────┴───────────────────────┤
+│ 10 rows 4 columns │
+└────────────────────────────────────────────────────────────────────────────────┘
+```
+RIGHT: Returns the right part of a character string with the specified number of characters.
+```python
+query = """SELECT FIRST, RIGHT(FIRST, 6) AS LastSixChars FROM patients LIMIT 10;
+"""
+
+conn.sql(query)
+```
+```python
+┌─────────────────┬──────────────┐
+│ FIRST │ LastSixChars │
+│ varchar │ varchar │
+├─────────────────┼──────────────┤
+│ Jacinto644 │ nto644 │
+│ Alva958 │ lva958 │
+│ Jimmie93 │ mmie93 │
+│ Gregorio366 │ rio366 │
+│ Karyn217 │ ryn217 │
+│ Jayson808 │ son808 │
+│ José Eduardo181 │ rdo181 │
+│ Milo271 │ ilo271 │
+│ Karyn217 │ ryn217 │
+│ Jeffrey461 │ rey461 │
+├─────────────────┴──────────────┤
+│ 10 rows 2 columns │
+└────────────────────────────────┘
+```
+SUBSTRING: Returns part of a character, binary, text, or image expression in SQL Server.
+```python
+query = """SELECT FIRST, SUBSTRING(FIRST, 1, 4) AS FirstFourChars FROM patients LIMIT 5;
+"""
+
+conn.sql(query)
+```
+```python
+┌─────────────┬────────────────┐
+│ FIRST │ FirstFourChars │
+│ varchar │ varchar │
+├─────────────┼────────────────┤
+│ Jacinto644 │ Jaci │
+│ Alva958 │ Alva │
+│ Jimmie93 │ Jimm │
+│ Gregorio366 │ Greg │
+│ Karyn217 │ Kary │
+└─────────────┴────────────────┘
+```
+REGEXP_REPLACE can use regular expressions to strip numeric characters:
+```python
+query = """SELECT
+FIRST, REGEXP_REPLACE(FIRST, '[0-9]', '', 'g') AS FirstNameStripped, LAST, REGEXP_REPLACE(LAST, '[0-9]', '', 'g') AS LastNameStripped
+FROM patients;
+"""
+
+conn.sql(query)
+```
+```python
+
+query = """SELECT
+FIRST, REGEXP_REPLACE(FIRST, '[0-9]', '', 'g') AS FirstNameStripped, LAST, REGEXP_REPLACE(LAST, '[0-9]', '', 'g') AS LastNameStripped
+FROM patients;
+"""
+
+conn.sql(query)
+┌─────────────────┬───────────────────┬────────────────┬──────────────────┐
+│ FIRST │ FirstNameStripped │ LAST │ LastNameStripped │
+│ varchar │ varchar │ varchar │ varchar │
+├─────────────────┼───────────────────┼────────────────┼──────────────────┤
+│ Jacinto644 │ Jacinto │ Kris249 │ Kris │
+│ Alva958 │ Alva │ Krajcik437 │ Krajcik │
+│ Jimmie93 │ Jimmie │ Harris789 │ Harris │
+│ Gregorio366 │ Gregorio │ Auer97 │ Auer │
+│ Karyn217 │ Karyn │ Mueller846 │ Mueller │
+│ Jayson808 │ Jayson │ Fadel536 │ Fadel │
+│ José Eduardo181 │ José Eduardo │ Gómez206 │ Gómez │
+│ Milo271 │ Milo │ Feil794 │ Feil │
+│ Karyn217 │ Karyn │ Metz686 │ Metz │
+│ Jeffrey461 │ Jeffrey │ Greenfelder433 │ Greenfelder │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ · │ · │ · │ · │
+│ Raymond398 │ Raymond │ Kuvalis369 │ Kuvalis │
+│ Gearldine455 │ Gearldine │ Boyer713 │ Boyer │
+│ Nichol11 │ Nichol │ Gleichner915 │ Gleichner │
+│ Louvenia131 │ Louvenia │ Marks830 │ Marks │
+│ Raymon366 │ Raymon │ Beer512 │ Beer │
+│ Camelia346 │ Camelia │ Stamm704 │ Stamm │
+│ William805 │ William │ Pacocha935 │ Pacocha │
+│ Guillermo498 │ Guillermo │ Téllez750 │ Téllez │
+│ Milton509 │ Milton │ Bailey598 │ Bailey │
+│ Cecilia788 │ Cecilia │ Wisozk929 │ Wisozk │
+├─────────────────┴───────────────────┴────────────────┴──────────────────┤
+│ ? rows (>9999 rows, 20 shown) 4 columns │
+└─────────────────────────────────────────────────────────────────────────┘
+```
+___
+___
+
+### Creating a database and adding data to tables
+
+### CREATE, ALTER, DROP, INSERT commands
+___
+First lets have a in-memory database
+```python
+newdb = duckdb.connect()
+```
+CREATE TABLE: Create a books table to mimic a library catalog system.
+```python
+query = """CREATE TABLE books (
+ book_id INTEGER PRIMARY KEY,
+ title VARCHAR,
+ author VARCHAR,
+ published_year INTEGER
+);
+"""
+
+newdb.sql(query)
+```
+The CREATE TABLE statement defines the schema for this table:
+This statement creates a table with columns for the book ID, title, author, and the year of publication. The book_id column is designated as the primary key, ensuring that each record in the table is unique.
+___
+check
+```python
+newdb.sql("SHOW TABLES;")
+```
+```python
+┌─────────┐
+│ name │
+│ varchar │
+├─────────┤
+│ books │
+└─────────┘
+```
+check data
+```python
+newdb.sql("DESCRIBE books;")
+```
+```python
+
+newdb.sql("DESCRIBE books;")
+┌────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ book_id │ INTEGER │ NO │ PRI │ NULL │ NULL │
+│ title │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ author │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ published_year │ INTEGER │ YES │ NULL │ NULL │ NULL │
+└────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┘
+```
+Alter Table: After creating the table, you might need to add more information. Use the ALTER TABLE statement to add a new column:
+```python
+query = """ALTER TABLE books ADD COLUMN genre VARCHAR;
+"""
+newdb.sql(query)
+# This adds a genre column to the books table, allowing you to categorize books.
+```
+```python
+newdb.sql("DESCRIBE books;")
+```
+```python
+
+newdb.sql("DESCRIBE books;")
+┌────────────────┬─────────────┬─────────┬─────────┬─────────┬───────┐
+│ column_name │ column_type │ null │ key │ default │ extra │
+│ varchar │ varchar │ varchar │ varchar │ varchar │ int32 │
+├────────────────┼─────────────┼─────────┼─────────┼─────────┼───────┤
+│ book_id │ INTEGER │ NO │ PRI │ NULL │ NULL │
+│ title │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ author │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+│ published_year │ INTEGER │ YES │ NULL │ NULL │ NULL │
+│ genre │ VARCHAR │ YES │ NULL │ NULL │ NULL │
+└────────────────┴─────────────┴─────────┴─────────┴─────────┴───────┘
+```
+let's add another table which we will DROP later;
+```python
+query = """CREATE TABLE authors (
+ author_id INTEGER PRIMARY KEY,
+ firstname VARCHAR,
+ lastname VARCHAR,
+ birth_year INTEGER
+);
+"""
+
+newdb.sql(query)
+```
+
+```python
+newdb.sql("SHOW TABLES;")
+```
+Now let's DROP the author table
+```python
+query = """DROP TABLE IF EXISTS authors;
+"""
+
+newdb.sql(query)
+```
+
+```python
+newdb.sql("SHOW TABLES;")
+```
+### Let's add some data now
+### INSERT To populate the books table with data. This inserts 10 records into the books table, providing a diverse set of examples.
+```python
+query = """INSERT INTO books (book_id, title, author, published_year, genre) VALUES
+(1, '1984', 'George Orwell', 1949, 'Dystopian'),
+(2, 'The Great Gatsby', 'F. Scott Fitzgerald', 1925, 'Classic'),
+(3, 'To Kill a Mockingbird', 'Harper Lee', 1960, 'Classic'),
+(4, 'Pride and Prejudice', 'Jane Austen', 1813, 'Romance'),
+(5, 'The Hobbit', 'J.R.R. Tolkien', 1937, 'Fantasy'),
+(6, 'The Catcher in the Rye', 'J.D. Salinger', 1951, 'Literary Fiction'),
+(7, 'Harry Potter and the Sorcerer’s Stone', 'J.K. Rowling', 1997, 'Fantasy'),
+(8, 'The Da Vinci Code', 'Dan Brown', 2003, 'Thriller'),
+(9, 'Sapiens: A Brief History of Humankind', 'Yuval Noah Harari', 2011, 'Non-fiction'),
+(10, 'Beloved', 'Toni Morrison', 1987, 'Historical Fiction');"""
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT title AS "Book Title", author AS "Author Name", Genre FROM books;
+"""
+
+newdb.sql(query)
+```
+
+```python
+┌───────────────────────────────────────┬─────────────────────┬────────────────────┐
+│ Book Title │ Author Name │ genre │
+│ varchar │ varchar │ varchar │
+├───────────────────────────────────────┼─────────────────────┼────────────────────┤
+│ 1984 │ George Orwell │ Dystopian │
+│ The Great Gatsby │ F. Scott Fitzgerald │ Classic │
+│ To Kill a Mockingbird │ Harper Lee │ Classic │
+│ Pride and Prejudice │ Jane Austen │ Romance │
+│ The Hobbit │ J.R.R. Tolkien │ Fantasy │
+│ The Catcher in the Rye │ J.D. Salinger │ Literary Fiction │
+│ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ Fantasy │
+│ The Da Vinci Code │ Dan Brown │ Thriller │
+│ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ Non-fiction │
+│ Beloved │ Toni Morrison │ Historical Fiction │
+├───────────────────────────────────────┴─────────────────────┴────────────────────┤
+│ 10 rows 3 columns │
+└──────────────────────────────────────────────────────────────────────────────────┘
+```
+### Updating Data: UPDATE
+This changes the genre of the book titled '1984' to 'Science Fiction'.
+```python
+query = """UPDATE books SET genre = 'Science Fiction' WHERE title = '1984';
+"""
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT title AS "Book Title", author AS "Author Name", Genre FROM books;
+"""
+
+newdb.sql(query)
+```
+
+```python
+┌───────────────────────────────────────┬─────────────────────┬────────────────────┐
+│ Book Title │ Author Name │ genre │
+│ varchar │ varchar │ varchar │
+├───────────────────────────────────────┼─────────────────────┼────────────────────┤
+│ 1984 │ George Orwell │ Science Fiction │
+│ The Great Gatsby │ F. Scott Fitzgerald │ Classic │
+│ To Kill a Mockingbird │ Harper Lee │ Classic │
+│ Pride and Prejudice │ Jane Austen │ Romance │
+│ The Hobbit │ J.R.R. Tolkien │ Fantasy │
+│ The Catcher in the Rye │ J.D. Salinger │ Literary Fiction │
+│ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ Fantasy │
+│ The Da Vinci Code │ Dan Brown │ Thriller │
+│ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ Non-fiction │
+│ Beloved │ Toni Morrison │ Historical Fiction │
+├───────────────────────────────────────┴─────────────────────┴────────────────────┤
+│ 10 rows 3 columns │
+└──────────────────────────────────────────────────────────────────────────────────┘
+```
+## DELETE
+e.g. To remove a book from the catalog
+
+```python
+query = """DELETE FROM books WHERE title = 'The Great Gatsby';"""
+newdb.sql(query)
+```
+
+```python
+query = """SELECT title AS "Book Title", author AS "Author Name", Genre FROM books;
+"""
+
+newdb.sql(query)
+```
+
+```python
+┌───────────────────────────────────────┬───────────────────┬────────────────────┐
+│ Book Title │ Author Name │ genre │
+│ varchar │ varchar │ varchar │
+├───────────────────────────────────────┼───────────────────┼────────────────────┤
+│ 1984 │ George Orwell │ Science Fiction │
+│ To Kill a Mockingbird │ Harper Lee │ Classic │
+│ Pride and Prejudice │ Jane Austen │ Romance │
+│ The Hobbit │ J.R.R. Tolkien │ Fantasy │
+│ The Catcher in the Rye │ J.D. Salinger │ Literary Fiction │
+│ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ Fantasy │
+│ The Da Vinci Code │ Dan Brown │ Thriller │
+│ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ Non-fiction │
+│ Beloved │ Toni Morrison │ Historical Fiction │
+└───────────────────────────────────────┴───────────────────┴────────────────────┘
+```
+### TRUNCATE
+- The TRUNCATE TABLE statement is used to delete all rows in a table without logging the individual row deletions.
+- This is faster than using DELETE without a WHERE clause because TRUNCATE immediately deallocates data pages used by the table.
+- Use TRUNCATE when you want to quickly remove all records from a table but keep the table structure for future use.
+- Unlike DELETE, TRUNCATE does not generate a large number of transaction logs, making it more efficient for completely clearing a table.
+
+```python
+query = """TRUNCATE TABLE books;"""
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT * FROM books;
+"""
+
+newdb.sql(query)
+```
+#### Adding one more table; named 'authors'
+```python
+query = """CREATE TABLE authors (
+ author_id INTEGER PRIMARY KEY,
+ name VARCHAR,
+ birth_year INTEGER,
+ nationality VARCHAR
+);
+"""
+
+newdb.sql(query)
+```
+Add information the authors table
+```python
+query = """INSERT INTO authors (author_id, name, birth_year, nationality) VALUES
+(1, 'George Orwell', 1903, 'British'),
+(2, 'F. Scott Fitzgerald', 1896, 'American'),
+(3, 'Harper Lee', 1926, 'American'),
+(4, 'Jane Austen', 1775, 'British'),
+(5, 'J.R.R. Tolkien', 1892, 'British'),
+(6, 'J.D. Salinger', 1919, 'American'),
+(7, 'J.K. Rowling', 1965, 'British'),
+(8, 'Dan Brown', 1964, 'American'),
+(9, 'Yuval Noah Harari', 1976, 'Israeli'),
+(10, 'Toni Morrison', 1931, 'American');
+"""
+
+
+newdb.sql(query)
+```
+
+```python
+query = """SELECT * FROM books;
+"""
+
+newdb.sql(query)
+```
+```python
+┌─────────┬───────────────────────────────────────┬─────────────────────┬────────────────┬────────────────────┐
+│ book_id │ title │ author │ published_year │ genre │
+│ int64 │ varchar │ varchar │ int64 │ varchar │
+├─────────┼───────────────────────────────────────┼─────────────────────┼────────────────┼────────────────────┤
+│ 1 │ 1984 │ George Orwell │ 1949 │ Dystopian │
+│ 2 │ The Great Gatsby │ F. Scott Fitzgerald │ 1925 │ Classic │
+│ 3 │ To Kill a Mockingbird │ Harper Lee │ 1960 │ Classic │
+│ 4 │ Pride and Prejudice │ Jane Austen │ 1813 │ Romance │
+│ 5 │ The Hobbit │ J.R.R. Tolkien │ 1937 │ Fantasy │
+│ 6 │ The Catcher in the Rye │ J.D. Salinger │ 1951 │ Literary Fiction │
+│ 7 │ Harry Potter and the Sorcerer’s Stone │ J.K. Rowling │ 1997 │ Fantasy │
+│ 8 │ The Da Vinci Code │ Dan Brown │ 2003 │ Thriller │
+│ 9 │ Sapiens: A Brief History of Humankind │ Yuval Noah Harari │ 2011 │ Non-fiction │
+│ 10 │ Beloved │ Toni Morrison │ 1987 │ Historical Fiction │
+├─────────┴───────────────────────────────────────┴─────────────────────┴────────────────┴────────────────────┤
+│ 10 rows 5 columns │
+└─────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
+```
+
+```python
+query = """SELECT * FROM authors;
+"""
+
+newdb.sql(query)
+```
+
+```python
+newdb.sql(query)
+┌───────────┬─────────────────────┬────────────┬─────────────┐
+│ author_id │ name │ birth_year │ nationality │
+│ int32 │ varchar │ int32 │ varchar │
+├───────────┼─────────────────────┼────────────┼─────────────┤
+│ 1 │ George Orwell │ 1903 │ British │
+│ 2 │ F. Scott Fitzgerald │ 1896 │ American │
+│ 3 │ Harper Lee │ 1926 │ American │
+│ 4 │ Jane Austen │ 1775 │ British │
+│ 5 │ J.R.R. Tolkien │ 1892 │ British │
+│ 6 │ J.D. Salinger │ 1919 │ American │
+│ 7 │ J.K. Rowling │ 1965 │ British │
+│ 8 │ Dan Brown │ 1964 │ American │
+│ 9 │ Yuval Noah Harari │ 1976 │ Israeli │
+│ 10 │ Toni Morrison │ 1931 │ American │
+├───────────┴─────────────────────┴────────────┴─────────────┤
+│ 10 rows 4 columns │
+└────────────────────────────────────────────────────────────┘
+```
+## EXPORTING Data to csv from SQL
+```python
+query = """EXPORT DATABASE './' (FORMAT CSV, DELIMITER ',');"""
+
+newdb.sql(query)
+```
+## Thank You!
diff --git a/11_sql_duckdb/index.html b/11_sql_duckdb/index.html
new file mode 100644
index 000000000..2a394295c
--- /dev/null
+++ b/11_sql_duckdb/index.html
@@ -0,0 +1,3487 @@
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ 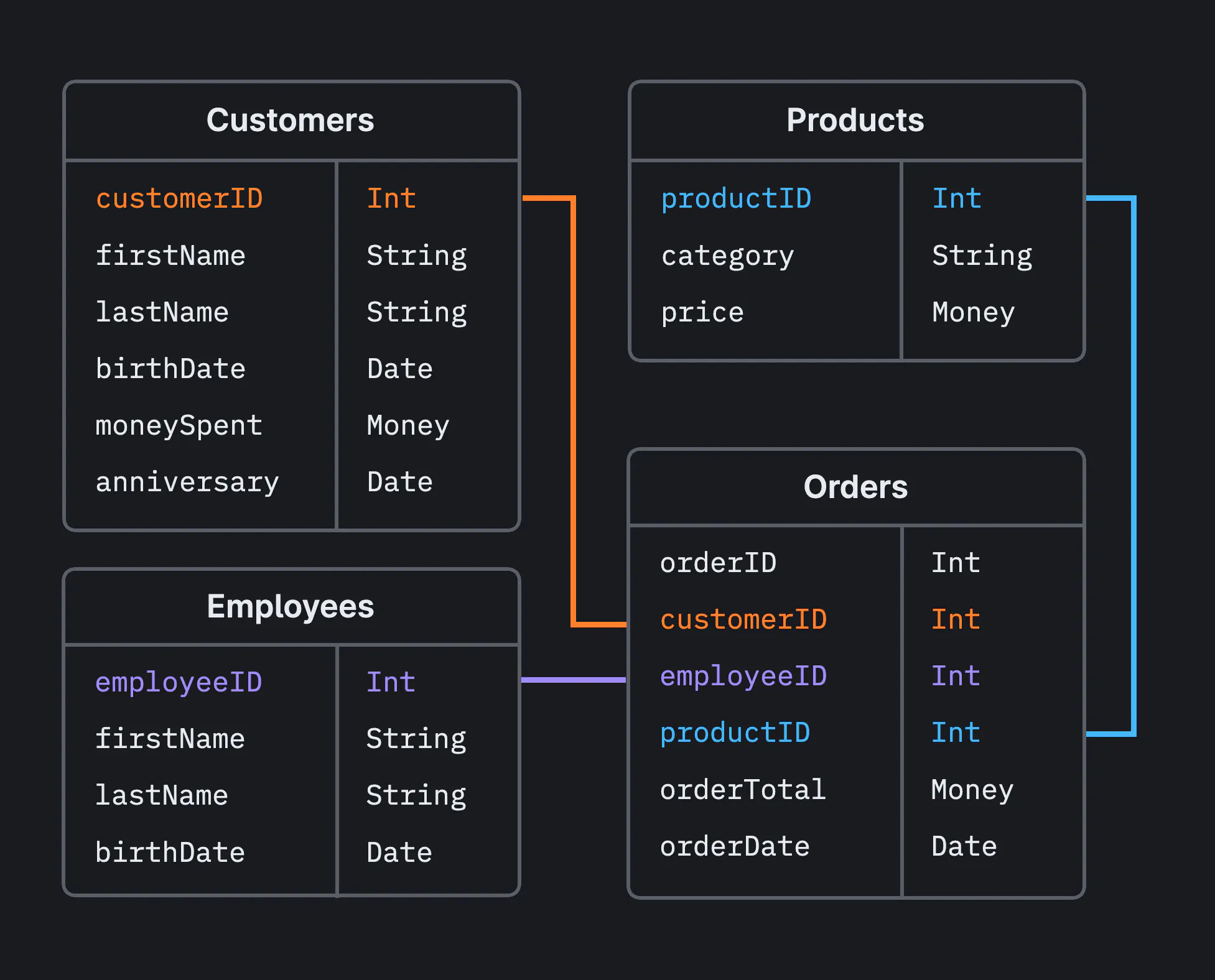
 + - credit: educba.com
+ - credit: educba.com