From 6733e79cf46372e80e119878f1bbeed5f5d142db Mon Sep 17 00:00:00 2001
From: Maarten Pronk <8655030+evetion@users.noreply.github.com>
Date: Tue, 27 Aug 2024 16:30:38 +0200
Subject: [PATCH 1/5] Add relationships between tables (#1756)
Fixes #1648
Fixes #1755 (by updating the edge style)
Fixes #1688 (node_id has to be unique)
Also sets the node_id and edge_id to be unique, so they are
autogenerated in the edit forms.
The current changes also will make the snapping + edge creation based on
coordinates obsolete, as the form now looks like this:
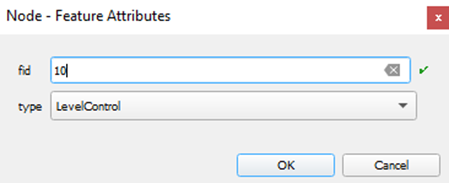
 There are buttons to open a form with the Node, a highlighter to blink
the node on the map, and a map selection panel, where one can click the
desired node.
---------
Co-authored-by: Martijn Visser
---
docs/dev/qgis_test_plan.qmd | 28 +++++---
docs/guide/qgis.qmd | 22 +++++--
python/ribasim/ribasim/styles/EdgeStyle.qml | 2 +-
ribasim_qgis/core/nodes.py | 20 +++++-
ribasim_qgis/widgets/dataset_widget.py | 73 ++++++++++++++++++++-
ribasim_qgis/widgets/nodes_widget.py | 2 +
ribasim_qgis/widgets/ribasim_widget.py | 6 +-
7 files changed, 135 insertions(+), 18 deletions(-)
diff --git a/docs/dev/qgis_test_plan.qmd b/docs/dev/qgis_test_plan.qmd
index 407dbdae1..5f4cee10d 100644
--- a/docs/dev/qgis_test_plan.qmd
+++ b/docs/dev/qgis_test_plan.qmd
@@ -29,7 +29,7 @@ Before starting with data, perform the following tests to see if the plugin does
- _In the Model tab, the "Add to QGIS" and "Remove from Dataset" buttons are disabled_.
- _In the Nodes tab, all buttons are disabled_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1678)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1678)
# New model tests
@@ -64,7 +64,7 @@ Before starting with data, perform the following tests to see if the plugin does
- Press OK: _An error is given that the database.gpkg already exists, test2.toml is not written_.
- Cleanup: Delete the created files from disk.
-[Failing](https://github.com/Deltares/Ribasim/issues/1679)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1679)
## New model with same name
@@ -83,7 +83,7 @@ Before starting with data, perform the following tests to see if the plugin does
Unsure if this is wanted behavior, as we said we wanted to overwrite the TOML file, and therefore also the database file.
Perhaps we should create folders instead of TOML files only.
-[Failing](https://github.com/Deltares/Ribasim/issues/1681)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1681)
## New model and clean new project cleans Ribasim plugin
@@ -95,7 +95,7 @@ Perhaps we should create folders instead of TOML files only.
- Press "Project > New": _Popup asks to save the project_.
- Press Discard: _Layers tab is emptied, Ribasim panel is emptied_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1682)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1682)
# Model tab button interaction tests
@@ -122,7 +122,7 @@ Intended behavior: The same model is loaded twice, but there is only a connectio
- Press "Add to QGIS": _Nothing happens_.
- Press the "Edge" layer in the Model tab: _"Add to QGIS" button is disabled_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1683)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1683)
## Remove from Dataset button
@@ -161,7 +161,7 @@ Perhaps better if we only allow removal of the optional tables. See this [bug re
- Go to the Nodes tab: _All buttons are enabled_.
- Press the Basin / time button: _A layer is added to the layers panel with that name, the button becomes disabled_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1685)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1685)
# Map interaction tests
@@ -199,10 +199,22 @@ See [issue](https://github.com/Deltares/Ribasim/issues/1688#issuecomment-2265315
- Select the Edge layer in the Layers tab: _edit buttons in the toolbar become enabled_.
- Enable snapping under View > Toolbars > Snapping Toolbars: _Magnet button is enabled and active_.
- Press the Add Line Feature button: _Mouse becomes a crosshair_.
-- Snap a line between the two nodes, click the two nodes and then right click to finish: _Popup shows with input, all is set to NULL_.
+- Snap a line between the two nodes, click the two nodes and then right click to finish: _Popup shows with input, most fields are set to NULL_.
- Press OK: _Line appears on screen between the two nodes_.
- Save the layer's edits: _The line becomes blue_.
-- Open the attribute table: _The information shows the from_node_id, to_node_id. This information matches the information from the Node table._
+- Open the attribute table: _The information shows the from\_node\_id, to\_node\_id. This information matches the information from the Node table._
+
+## Node selection on map triggers table selection
+
+- Open QGIS and ensure that the Ribasim plugin is installed and enabled.
+- Open the application via the Ribasim button on the QGIS toolbar: _Ribasim panel opens_.
+- Press the "Open" button in the Model tab: _file navigation window pops up_.
+- Choose an existing model from the `generated_testmodels` folder.
+- Press OK: _The model layers appear in the layer panel and on the map_.
+- Select the node layer, and make a subselection of nodes on the map: _Nodes are highlighted in yellow, including their edges_.
+- Open the Edge attribute table: _The highlighted rows are those with a from/to node\_id that was selected_.
+- Open any non-spatial attribute table: _The highlighted rows are those with an node\_id that was selected_.
+
# Tables tests
diff --git a/docs/guide/qgis.qmd b/docs/guide/qgis.qmd
index 5b3d5f97d..94abdee02 100644
--- a/docs/guide/qgis.qmd
+++ b/docs/guide/qgis.qmd
@@ -36,7 +36,7 @@ Turn on the edit mode to be able to add nodes on the map.
Add nodes to the map with a left click and select the node type.
-{fig-align="left"}
+
There are buttons to open a form with the Node, a highlighter to blink
the node on the map, and a map selection panel, where one can click the
desired node.
---------
Co-authored-by: Martijn Visser
---
docs/dev/qgis_test_plan.qmd | 28 +++++---
docs/guide/qgis.qmd | 22 +++++--
python/ribasim/ribasim/styles/EdgeStyle.qml | 2 +-
ribasim_qgis/core/nodes.py | 20 +++++-
ribasim_qgis/widgets/dataset_widget.py | 73 ++++++++++++++++++++-
ribasim_qgis/widgets/nodes_widget.py | 2 +
ribasim_qgis/widgets/ribasim_widget.py | 6 +-
7 files changed, 135 insertions(+), 18 deletions(-)
diff --git a/docs/dev/qgis_test_plan.qmd b/docs/dev/qgis_test_plan.qmd
index 407dbdae1..5f4cee10d 100644
--- a/docs/dev/qgis_test_plan.qmd
+++ b/docs/dev/qgis_test_plan.qmd
@@ -29,7 +29,7 @@ Before starting with data, perform the following tests to see if the plugin does
- _In the Model tab, the "Add to QGIS" and "Remove from Dataset" buttons are disabled_.
- _In the Nodes tab, all buttons are disabled_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1678)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1678)
# New model tests
@@ -64,7 +64,7 @@ Before starting with data, perform the following tests to see if the plugin does
- Press OK: _An error is given that the database.gpkg already exists, test2.toml is not written_.
- Cleanup: Delete the created files from disk.
-[Failing](https://github.com/Deltares/Ribasim/issues/1679)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1679)
## New model with same name
@@ -83,7 +83,7 @@ Before starting with data, perform the following tests to see if the plugin does
Unsure if this is wanted behavior, as we said we wanted to overwrite the TOML file, and therefore also the database file.
Perhaps we should create folders instead of TOML files only.
-[Failing](https://github.com/Deltares/Ribasim/issues/1681)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1681)
## New model and clean new project cleans Ribasim plugin
@@ -95,7 +95,7 @@ Perhaps we should create folders instead of TOML files only.
- Press "Project > New": _Popup asks to save the project_.
- Press Discard: _Layers tab is emptied, Ribasim panel is emptied_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1682)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1682)
# Model tab button interaction tests
@@ -122,7 +122,7 @@ Intended behavior: The same model is loaded twice, but there is only a connectio
- Press "Add to QGIS": _Nothing happens_.
- Press the "Edge" layer in the Model tab: _"Add to QGIS" button is disabled_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1683)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1683)
## Remove from Dataset button
@@ -161,7 +161,7 @@ Perhaps better if we only allow removal of the optional tables. See this [bug re
- Go to the Nodes tab: _All buttons are enabled_.
- Press the Basin / time button: _A layer is added to the layers panel with that name, the button becomes disabled_.
-[Failing](https://github.com/Deltares/Ribasim/issues/1685)
+❌ [Failing](https://github.com/Deltares/Ribasim/issues/1685)
# Map interaction tests
@@ -199,10 +199,22 @@ See [issue](https://github.com/Deltares/Ribasim/issues/1688#issuecomment-2265315
- Select the Edge layer in the Layers tab: _edit buttons in the toolbar become enabled_.
- Enable snapping under View > Toolbars > Snapping Toolbars: _Magnet button is enabled and active_.
- Press the Add Line Feature button: _Mouse becomes a crosshair_.
-- Snap a line between the two nodes, click the two nodes and then right click to finish: _Popup shows with input, all is set to NULL_.
+- Snap a line between the two nodes, click the two nodes and then right click to finish: _Popup shows with input, most fields are set to NULL_.
- Press OK: _Line appears on screen between the two nodes_.
- Save the layer's edits: _The line becomes blue_.
-- Open the attribute table: _The information shows the from_node_id, to_node_id. This information matches the information from the Node table._
+- Open the attribute table: _The information shows the from\_node\_id, to\_node\_id. This information matches the information from the Node table._
+
+## Node selection on map triggers table selection
+
+- Open QGIS and ensure that the Ribasim plugin is installed and enabled.
+- Open the application via the Ribasim button on the QGIS toolbar: _Ribasim panel opens_.
+- Press the "Open" button in the Model tab: _file navigation window pops up_.
+- Choose an existing model from the `generated_testmodels` folder.
+- Press OK: _The model layers appear in the layer panel and on the map_.
+- Select the node layer, and make a subselection of nodes on the map: _Nodes are highlighted in yellow, including their edges_.
+- Open the Edge attribute table: _The highlighted rows are those with a from/to node\_id that was selected_.
+- Open any non-spatial attribute table: _The highlighted rows are those with an node\_id that was selected_.
+
# Tables tests
diff --git a/docs/guide/qgis.qmd b/docs/guide/qgis.qmd
index 5b3d5f97d..94abdee02 100644
--- a/docs/guide/qgis.qmd
+++ b/docs/guide/qgis.qmd
@@ -36,7 +36,7 @@ Turn on the edit mode to be able to add nodes on the map.
Add nodes to the map with a left click and select the node type.
-{fig-align="left"}
+ Turn the edit mode off and save the edits to the Nodes layer.
@@ -53,7 +53,8 @@ A record can be selected by clicking on the row number.
{fig-align="left"}
-Adjust the content. If you prefer, it also works to copy data with the same columns from
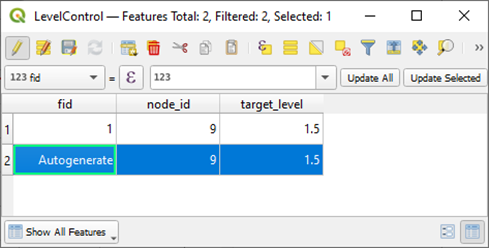
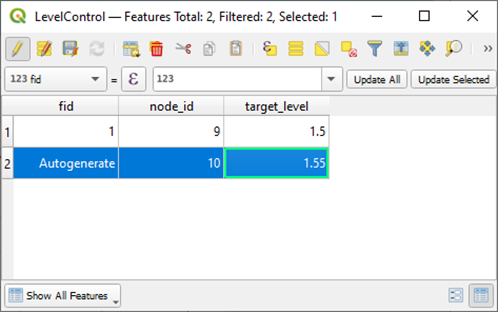
+Adjust the content. Note that the `node_id` field is connected to the Node layer and thus must be set to an existing `node_id`.
+If you prefer, it also works to copy data with the same columns from
Excel. Turn off edit mode and save changes to the layer.
{fig-align="left"}
@@ -63,7 +64,7 @@ Excel. Turn off edit mode and save changes to the layer.
### Turn on snapping
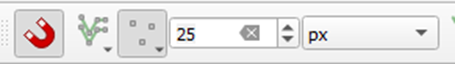
Make sure the Snapping Toolbar is visible, by going to the View > Toolbars menu. Turn on
-snapping mode by clicking the magnet and set the snapping distance to 25 pixels.
+snapping mode by clicking the magnet and set the snapping distance to 25 pixels. The keyboard shortcut for snapping is `s` (once the toolbar is enabled).
{fig-align="left"}
@@ -79,7 +80,11 @@ Select "Add line feature".
Create a connection by left clicking a source node and right clicking the destination node.
-{fig-align="left"}
+
Turn the edit mode off and save the edits to the Nodes layer.
@@ -53,7 +53,8 @@ A record can be selected by clicking on the row number.
{fig-align="left"}
-Adjust the content. If you prefer, it also works to copy data with the same columns from
+Adjust the content. Note that the `node_id` field is connected to the Node layer and thus must be set to an existing `node_id`.
+If you prefer, it also works to copy data with the same columns from
Excel. Turn off edit mode and save changes to the layer.
{fig-align="left"}
@@ -63,7 +64,7 @@ Excel. Turn off edit mode and save changes to the layer.
### Turn on snapping
Make sure the Snapping Toolbar is visible, by going to the View > Toolbars menu. Turn on
-snapping mode by clicking the magnet and set the snapping distance to 25 pixels.
+snapping mode by clicking the magnet and set the snapping distance to 25 pixels. The keyboard shortcut for snapping is `s` (once the toolbar is enabled).
{fig-align="left"}
@@ -79,7 +80,11 @@ Select "Add line feature".
Create a connection by left clicking a source node and right clicking the destination node.
-{fig-align="left"}
+ +
+A form where one can change the edge attributes will pop up. Once done with editing, click ok.
+
+
+
+A form where one can change the edge attributes will pop up. Once done with editing, click ok.
+
+ Now leave the edit mode and save the results to the layer. Your model is now ready to run. See
@@ -91,6 +96,15 @@ Now leave the edit mode and save the results to the layer. Your model is now rea
- In your model directory there is now a `results/` folder with `basin.arrow` and
`flow.arrow` output files.
+# Inspect a (large) model {#sec-inspect}
+
+For larger models the node tables can grow quite large. To facilitate inspection,
+the tables are linked via the `node_id` field to the Node table, and react to the selection of the Node layer. That is, on selection of certain nodes---either via the map or the attribute table---the selection is also made in all related tables. This is also the case for the Edge layer.
+It helps to set the attribute table of a table of interest to show selected features only (using the dropdown button on the bottom left).
+
+
Now leave the edit mode and save the results to the layer. Your model is now ready to run. See
@@ -91,6 +96,15 @@ Now leave the edit mode and save the results to the layer. Your model is now rea
- In your model directory there is now a `results/` folder with `basin.arrow` and
`flow.arrow` output files.
+# Inspect a (large) model {#sec-inspect}
+
+For larger models the node tables can grow quite large. To facilitate inspection,
+the tables are linked via the `node_id` field to the Node table, and react to the selection of the Node layer. That is, on selection of certain nodes---either via the map or the attribute table---the selection is also made in all related tables. This is also the case for the Edge layer.
+It helps to set the attribute table of a table of interest to show selected features only (using the dropdown button on the bottom left).
+
+ +
+
# Inspecting results {#sec-results}
Before inspecting the results, verify that the run was successfully and the output files are there.
diff --git a/python/ribasim/ribasim/styles/EdgeStyle.qml b/python/ribasim/ribasim/styles/EdgeStyle.qml
index acbb8381b..5b9bee800 100644
--- a/python/ribasim/ribasim/styles/EdgeStyle.qml
+++ b/python/ribasim/ribasim/styles/EdgeStyle.qml
@@ -555,7 +555,7 @@
-
+
diff --git a/ribasim_qgis/core/nodes.py b/ribasim_qgis/core/nodes.py
index f257fe8ee..38fbbeee7 100644
--- a/ribasim_qgis/core/nodes.py
+++ b/ribasim_qgis/core/nodes.py
@@ -123,6 +123,15 @@ def set_dropdown(self, name: str, options: set[str]) -> None:
)
layer.setEditorWidgetSetup(index, setup)
+ def set_unique(self, name: str) -> None:
+ layer = self.layer
+ index = layer.fields().indexFromName(name)
+ setup = QgsEditorWidgetSetup(
+ "UniqueValues",
+ {},
+ )
+ layer.setEditorWidgetSetup(index, setup)
+
def set_read_only(self) -> None:
pass
@@ -134,8 +143,11 @@ def layer_from_geopackage(self) -> QgsVectorLayer:
self.layer = QgsVectorLayer(
f"{self._path}|layername={self.input_type()}", self.input_type()
)
- # Load style from database if exists
- self.layer.loadDefaultStyle()
+ # Load style from database if exists, otherwise load and save default qml style
+ _, success = self.layer.loadDefaultStyle()
+ if not success:
+ self.load_default_style()
+ self.save_style()
# Connect signal to save style to database when changed
self.layer.styleChanged.connect(self.save_style)
return self.layer
@@ -214,6 +226,7 @@ def set_editor_widget(self) -> None:
layer = self.layer
node_type_field_index = layer.fields().indexFromName("node_type")
self.set_dropdown("node_type", NONSPATIALNODETYPES)
+ self.set_unique("node_id")
layer_form_config = layer.editFormConfig()
layer_form_config.setReuseLastValue(node_type_field_index, True)
@@ -267,6 +280,7 @@ def set_editor_widget(self) -> None:
layer = self.layer
self.set_dropdown("edge_type", EDGETYPES)
+ self.set_unique("edge_id")
layer_form_config = layer.editFormConfig()
layer.setEditFormConfig(layer_form_config)
@@ -859,7 +873,7 @@ def attributes(cls) -> list[QgsField]:
}
NONSPATIALNODETYPES: set[str] = {
cls.nodetype() for cls in Input.__subclasses__() if not cls.is_spatial()
-}
+} | {"Terminal"}
EDGETYPES = {"flow", "control"}
SPATIALCONTROLNODETYPES = {
"ContinuousControl",
diff --git a/ribasim_qgis/widgets/dataset_widget.py b/ribasim_qgis/widgets/dataset_widget.py
index d43eff23a..b84656e16 100644
--- a/ribasim_qgis/widgets/dataset_widget.py
+++ b/ribasim_qgis/widgets/dataset_widget.py
@@ -8,6 +8,7 @@
from __future__ import annotations
from datetime import datetime
+from functools import partial
from pathlib import Path
from typing import Any, cast
@@ -27,8 +28,11 @@
QWidget,
)
from qgis.core import (
+ QgsEditorWidgetSetup,
+ QgsFeatureRequest,
QgsMapLayer,
QgsProject,
+ QgsRelation,
QgsVectorLayer,
)
@@ -208,6 +212,32 @@ def add_selection_to_qgis(self) -> None:
for item in selection:
self.add_item_to_qgis(item)
+ @staticmethod
+ def add_relationship(from_layer, to_layer_id, name, fk="node_id") -> None:
+ rel = QgsRelation()
+ rel.setReferencingLayer(from_layer.id())
+ rel.setReferencedLayer(to_layer_id)
+ rel.setName(name)
+ rel.setStrength(rel.RelationStrength.Composition) # type: ignore
+ rel.addFieldPair(fk, "node_id")
+ rel.generateId()
+ instance = QgsProject.instance()
+ assert instance is not None
+ rel_manager = instance.relationManager()
+ assert rel_manager is not None
+ rel_manager.addRelation(rel)
+
+ # Also use the relationship as an editor widget
+ field_index = from_layer.fields().indexFromName(fk)
+ setup = QgsEditorWidgetSetup(
+ "RelationReference",
+ {
+ "Relation": rel.id(),
+ "MapIdentification": True,
+ },
+ )
+ from_layer.setEditorWidgetSetup(field_index, setup)
+
def load_geopackage(self) -> None:
"""Load the layers of a GeoPackage into the Layers Panel"""
self.dataset_tree.clear()
@@ -221,25 +251,66 @@ def load_geopackage(self) -> None:
node = nodes.pop("Node")
item = self.dataset_tree.add_node_layer(node)
self.add_item_to_qgis(item)
+ # Make sure node_id shows up in relationships
+ node.layer.setDisplayExpression("node_id")
edge = nodes.pop("Edge")
item = self.dataset_tree.add_node_layer(edge)
self.add_item_to_qgis(item)
+ self.add_relationship(
+ edge.layer, node.layer.id(), "EdgeFromNode", "from_node_id"
+ )
+ self.add_relationship(edge.layer, node.layer.id(), "EdgeToNode", "to_node_id")
basin_area_layer = nodes.pop("Basin / area", None)
if basin_area_layer is not None:
item = self.dataset_tree.add_node_layer(basin_area_layer)
self.add_item_to_qgis(item)
+ self.add_relationship(
+ basin_area_layer.layer, node.layer.id(), "Basin / area"
+ )
# Add the remaining layers
- for node_layer in nodes.values():
+ for table_name, node_layer in nodes.items():
item = self.dataset_tree.add_node_layer(node_layer)
self.add_item_to_qgis(item)
+ self.add_relationship(node_layer.layer, node.layer.id(), table_name)
# Connect node and edge layer to derive connectivities.
self.node_layer = node.layer
+ assert self.node_layer is not None
self.edge_layer = edge.layer
self.edge_layer.editingStopped.connect(self.connect_nodes)
+
+ def filterbyrel(relationships, feature_ids):
+ """Filter all related tables by the selected features in the node table."""
+ ids = []
+ selection = QgsFeatureRequest().setFilterFids(feature_ids)
+ for rel in relationships:
+ for feature in rel.referencedLayer().getFeatures(selection):
+ ids.extend(f.id() for f in rel.getRelatedFeatures(feature))
+
+ rel.referencingLayer().selectByIds(ids)
+
+ # When the Node selection changes, filter all related tables
+ edge_rels = []
+ instance = QgsProject.instance()
+ assert instance is not None
+ rel_manager = instance.relationManager()
+ assert rel_manager is not None
+ for rel in rel_manager.relations().values():
+ # Edge relations are special, they have two references to the Node table
+ referencing = rel.referencingLayer()
+ referenced = rel.referencedLayer()
+ assert referencing is not None
+ assert referenced is not None
+
+ if referencing.name() == "Edge":
+ edge_rels.append(rel)
+ else:
+ referenced.selectionChanged.connect(partial(filterbyrel, [rel]))
+
+ self.node_layer.selectionChanged.connect(partial(filterbyrel, edge_rels))
return
def new_model(self) -> None:
diff --git a/ribasim_qgis/widgets/nodes_widget.py b/ribasim_qgis/widgets/nodes_widget.py
index 689e0e78a..cdaa293ae 100644
--- a/ribasim_qgis/widgets/nodes_widget.py
+++ b/ribasim_qgis/widgets/nodes_widget.py
@@ -67,5 +67,7 @@ def new_node_layer(self, node_type: str) -> None:
node.write()
# Add to QGIS
self.ribasim_widget.add_layer(node.layer, "Ribasim Input", labels=node.labels)
+ # Setup relationship
+ self.ribasim_widget.add_relationship(node.layer, node_type)
# Add to dataset tree
self.ribasim_widget.add_node_layer(node)
diff --git a/ribasim_qgis/widgets/ribasim_widget.py b/ribasim_qgis/widgets/ribasim_widget.py
index 2635865e8..8af4689c4 100644
--- a/ribasim_qgis/widgets/ribasim_widget.py
+++ b/ribasim_qgis/widgets/ribasim_widget.py
@@ -106,7 +106,7 @@ def create_groups(self, name: str) -> None:
assert project is not None
root = project.layerTreeRoot()
assert root is not None
- self.group = root.addGroup(name)
+ self.group = root.insertGroup(0, name) # insert at the top
self.create_subgroup(name, "Ribasim Input")
def add_to_group(self, maplayer: Any, destination: str, on_top: bool):
@@ -182,3 +182,7 @@ def add_layer(
self.add_to_group(maplayer, destination, on_top)
return maplayer
+
+ def add_relationship(self, new_layer, name: str) -> None:
+ assert self.node_layer is not None
+ self.__dataset_widget.add_relationship(new_layer, self.node_layer.id(), name)
From 4cf93b5ec7e38135f4e870d0bb714d0a06890068 Mon Sep 17 00:00:00 2001
From: Bart de Koning <74617371+SouthEndMusic@users.noreply.github.com>
Date: Tue, 27 Aug 2024 18:43:58 +0200
Subject: [PATCH 2/5] Give no bottleneck warning if no bottleneck basins were
found (#1770)
Fixes https://github.com/Deltares/Ribasim/issues/1759
---
core/src/logging.jl | 4 +++-
1 file changed, 3 insertions(+), 1 deletion(-)
diff --git a/core/src/logging.jl b/core/src/logging.jl
index 18f474de9..13f3dc020 100644
--- a/core/src/logging.jl
+++ b/core/src/logging.jl
@@ -46,7 +46,9 @@ function log_bottlenecks(model; converged::Bool)
end
push!(errors, node_id => error)
end
- @logmsg level "Convergence bottlenecks in descending order of severity:" errors...
+ if !isempty(errors)
+ @logmsg level "Convergence bottlenecks in descending order of severity:" errors...
+ end
else
algorithm = model.config.solver.algorithm
@logmsg level "Convergence bottlenecks are not shown for the chosen solver algorithm." algorithm
From 7ec16c76f0ea3931e703cee26cd4b80e92fc0152 Mon Sep 17 00:00:00 2001
From: Maarten Pronk
Date: Thu, 29 Aug 2024 13:43:48 +0200
Subject: [PATCH 3/5] Error when the toml or database can't be found. (#1773)
Fixes #1652
---
python/ribasim/ribasim/model.py | 9 ++++++++-
python/ribasim/tests/test_io.py | 1 +
python/ribasim/tests/test_model.py | 17 +++++++++++++++++
3 files changed, 26 insertions(+), 1 deletion(-)
diff --git a/python/ribasim/ribasim/model.py b/python/ribasim/ribasim/model.py
index 11c50da96..947738e80 100644
--- a/python/ribasim/ribasim/model.py
+++ b/python/ribasim/ribasim/model.py
@@ -253,6 +253,8 @@ def read(cls, filepath: str | PathLike[str]) -> "Model":
filepath : str | PathLike[str]
The path to the TOML file.
"""
+ if not Path(filepath).is_file():
+ raise FileNotFoundError(f"File '{filepath}' does not exist.")
return cls(filepath=filepath) # type: ignore
def write(self, filepath: str | PathLike[str]) -> Path:
@@ -290,7 +292,12 @@ def _load(cls, filepath: Path | None) -> dict[str, Any]:
directory = filepath.parent / config.get("input_dir", ".")
context_file_loading.get()["directory"] = directory
- context_file_loading.get()["database"] = directory / "database.gpkg"
+ db_path = directory / "database.gpkg"
+
+ if not db_path.is_file():
+ raise FileNotFoundError(f"Database file '{db_path}' does not exist.")
+
+ context_file_loading.get()["database"] = db_path
return config
else:
diff --git a/python/ribasim/tests/test_io.py b/python/ribasim/tests/test_io.py
index 60d977316..5aedd70da 100644
--- a/python/ribasim/tests/test_io.py
+++ b/python/ribasim/tests/test_io.py
@@ -299,5 +299,6 @@ def test_datetime_timezone():
def test_minimal_toml():
# Check if the TOML used in QGIS tests is still valid.
toml_path = Path(__file__).parents[3] / "ribasim_qgis/tests/data/simple_valid.toml"
+ (toml_path.parent / "database.gpkg").touch() # database file must exist for `read`
model = ribasim.Model.read(toml_path)
assert model.crs == "EPSG:28992"
diff --git a/python/ribasim/tests/test_model.py b/python/ribasim/tests/test_model.py
index 6b96988c9..7f2dfdee5 100644
--- a/python/ribasim/tests/test_model.py
+++ b/python/ribasim/tests/test_model.py
@@ -4,6 +4,7 @@
import numpy as np
import pandas as pd
import pytest
+import tomli_w
import xugrid
from pydantic import ValidationError
from pyproj import CRS
@@ -228,3 +229,19 @@ def test_styles(tabulated_rating_curve: Model, tmp_path):
model.write(tmp_path / "basic" / "ribasim.toml")
with connect(tmp_path / "basic" / "database.gpkg") as conn:
assert conn.execute("SELECT COUNT(*) FROM layer_styles").fetchone()[0] == 3
+
+
+def test_non_existent_files(tmp_path):
+ with pytest.raises(
+ FileNotFoundError, match="File 'non_existent_file.toml' does not exist."
+ ):
+ Model.read("non_existent_file.toml")
+
+ # Create a TOML file without a database.gpkg
+ content = {"input_path": str(tmp_path)}
+ toml_path = tmp_path / "test.toml"
+ with open(toml_path, "wb") as f:
+ tomli_w.dump(content, f)
+
+ with pytest.raises(FileNotFoundError, match=r"Database file .* does not exist\."):
+ Model.read(toml_path)
From b80a79addac96c8ad1c7f4657af871da3c69d627 Mon Sep 17 00:00:00 2001
From: Maarten Pronk
Date: Thu, 29 Aug 2024 13:46:53 +0200
Subject: [PATCH 4/5] Enable Docker for QGIS tests (once more) (#1774)
Fixes #1751
Changes since last time:
- Also includes coverage
- Also runs other QGIS tests
---
.docker/compose.yml | 12 ++++++
.docker/start.sh | 7 ++++
.docker/stop.sh | 6 +++
.docker/test.sh | 4 ++
.github/workflows/qgis.yml | 40 ++++++++-----------
pixi.toml | 5 ++-
ribasim_qgis/.coveragerc | 8 ++--
ribasim_qgis/scripts/run_qgis_ui_tests.py | 2 +-
ribasim_qgis/tests/__init__.py | 20 ++++++++++
ribasim_qgis/tests/ui/__init__.py | 0
.../ui}/test_load_plugin.py | 0
ribasim_qgis/ui_tests/__init__.py | 9 -----
12 files changed, 74 insertions(+), 39 deletions(-)
create mode 100644 .docker/compose.yml
create mode 100755 .docker/start.sh
create mode 100755 .docker/stop.sh
create mode 100755 .docker/test.sh
create mode 100644 ribasim_qgis/tests/__init__.py
create mode 100644 ribasim_qgis/tests/ui/__init__.py
rename ribasim_qgis/{ui_tests => tests/ui}/test_load_plugin.py (100%)
delete mode 100644 ribasim_qgis/ui_tests/__init__.py
diff --git a/.docker/compose.yml b/.docker/compose.yml
new file mode 100644
index 000000000..8f6f42924
--- /dev/null
+++ b/.docker/compose.yml
@@ -0,0 +1,12 @@
+version: '2'
+
+services:
+ qgis:
+ image: qgis/qgis:release-3_34
+ container_name: qgis
+ volumes:
+ - ../ribasim_qgis/:/tests_directory/ribasim_qgis
+ environment:
+ - CI=true
+ - DISPLAY=:99
+ tty: true
diff --git a/.docker/start.sh b/.docker/start.sh
new file mode 100755
index 000000000..6c583b16b
--- /dev/null
+++ b/.docker/start.sh
@@ -0,0 +1,7 @@
+#!/usr/bin/env bash
+set -eux
+
+docker compose -f compose.yml up -d --force-recreate --remove-orphans
+echo "Installation of the plugin Ribasim"
+docker exec -t qgis sh -c "qgis_setup.sh ribasim_qgis"
+echo "Containers are running"
diff --git a/.docker/stop.sh b/.docker/stop.sh
new file mode 100755
index 000000000..08afc0ec6
--- /dev/null
+++ b/.docker/stop.sh
@@ -0,0 +1,6 @@
+#!/usr/bin/env bash
+set -eux
+
+echo 'Stopping/killing containers'
+docker compose -f compose.yml kill
+docker compose -f compose.yml rm -f
diff --git a/.docker/test.sh b/.docker/test.sh
new file mode 100755
index 000000000..b1ce0ec3b
--- /dev/null
+++ b/.docker/test.sh
@@ -0,0 +1,4 @@
+#!/usr/bin/env bash
+set -eux
+
+docker exec -t qgis sh -c "cd /tests_directory && xvfb-run -a qgis_testrunner.sh ribasim_qgis.tests"
diff --git a/.github/workflows/qgis.yml b/.github/workflows/qgis.yml
index fc9b302de..3275c8db4 100644
--- a/.github/workflows/qgis.yml
+++ b/.github/workflows/qgis.yml
@@ -8,27 +8,21 @@ on:
pull_request:
paths-ignore: [".teamcity/**"]
merge_group:
-concurrency:
- group: ${{ github.workflow }}-${{ github.ref }}
- cancel-in-progress: true
jobs:
- test:
- name: QGIS plugin ${{ matrix.os }}
- runs-on: ${{ matrix.os }}
- strategy:
- matrix:
- os:
- - ubuntu-latest
- - macOS-latest
- - windows-latest
- steps:
- - uses: actions/checkout@v4
- - uses: prefix-dev/setup-pixi@v0.8.1
- with:
- pixi-version: "latest"
- - name: Run tests
- run: pixi run test-ribasim-qgis-cov
- - name: Upload coverage to Codecov
- uses: codecov/codecov-action@v4
- with:
- token: ${{ secrets.CODECOV_TOKEN }}
+ test-qgis:
+ name: "Test"
+ runs-on: ubuntu-latest
+ defaults:
+ run:
+ working-directory: .docker
+ steps:
+ - uses: actions/checkout@v4
+ - uses: prefix-dev/setup-pixi@v0.8.1
+ with:
+ pixi-version: "latest"
+ - name: Run tests
+ run: pixi run test-ribasim-qgis-docker
+ - name: Upload coverage to Codecov
+ uses: codecov/codecov-action@v4
+ with:
+ token: ${{ secrets.CODECOV_TOKEN }}
diff --git a/pixi.toml b/pixi.toml
index e16056b63..346715472 100644
--- a/pixi.toml
+++ b/pixi.toml
@@ -128,6 +128,7 @@ install-ribasim-qgis = "python ribasim_qgis/scripts/install_ribasim_qgis.py"
install-imod-qgis = "python ribasim_qgis/scripts/install_qgis_plugin.py iMOD && python ribasim_qgis/scripts/enable_plugin.py imodqgis"
install-plugin-reloader-qgis = "python ribasim_qgis/scripts/install_qgis_plugin.py \"Plugin Reloader\" && python ribasim_qgis/scripts/enable_plugin.py plugin_reloader"
install-debugvs-qgis = "python ribasim_qgis/scripts/install_qgis_plugin.py debugvs==0.7 && python ribasim_qgis/scripts/enable_plugin.py debug_vs"
+test-ribasim-qgis-docker = { cmd = "sh ./start.sh; sh ./test.sh; sh ./stop.sh", cwd = ".docker" }
install-qgis-plugins = { depends_on = [
"install-plugin-reloader-qgis",
"install-ribasim-qgis",
@@ -137,10 +138,10 @@ install-qgis-plugins = { depends_on = [
test-ribasim-qgis-ui = { cmd = "python ribasim_qgis/scripts/run_qgis_ui_tests.py", depends_on = [
"install-ribasim-qgis",
] }
-test-ribasim-qgis = { cmd = "pytest --numprocesses=auto ribasim_qgis/tests", depends_on = [
+test-ribasim-qgis = { cmd = "pytest --numprocesses=auto ribasim_qgis/tests/core", depends_on = [
"install-ribasim-qgis",
] }
-test-ribasim-qgis-cov = { cmd = "pytest --numprocesses=auto --cov=ribasim_qgis --cov-report=xml --cov-config=ribasim_qgis/.coveragerc ribasim_qgis/tests", depends_on = [
+test-ribasim-qgis-cov = { cmd = "pytest --numprocesses=auto --cov=ribasim_qgis --cov-report=xml --cov-config=ribasim_qgis/.coveragerc ribasim_qgis/tests/core", depends_on = [
"install-ribasim-qgis",
] }
mypy-ribasim-qgis = "mypy ribasim_qgis"
diff --git a/ribasim_qgis/.coveragerc b/ribasim_qgis/.coveragerc
index 011bc9fa5..e14c37159 100644
--- a/ribasim_qgis/.coveragerc
+++ b/ribasim_qgis/.coveragerc
@@ -1,6 +1,6 @@
[run]
+source = .
omit =
- ribasim_qgis/resources.py
- ribasim_qgis/tests/*
- ribasim_qgis/tomllib/*
- ribasim_qgis/ui_tests/*
+ */resources.py
+ */tests/*
+ */tomllib/*
diff --git a/ribasim_qgis/scripts/run_qgis_ui_tests.py b/ribasim_qgis/scripts/run_qgis_ui_tests.py
index bee8e1096..04fda7c4d 100644
--- a/ribasim_qgis/scripts/run_qgis_ui_tests.py
+++ b/ribasim_qgis/scripts/run_qgis_ui_tests.py
@@ -9,7 +9,7 @@
"--nologo",
"--code",
"ribasim_qgis/scripts/qgis_testrunner.py",
- "ribasim_qgis.ui_tests",
+ "ribasim_qgis.tests",
],
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
diff --git a/ribasim_qgis/tests/__init__.py b/ribasim_qgis/tests/__init__.py
new file mode 100644
index 000000000..7034cc660
--- /dev/null
+++ b/ribasim_qgis/tests/__init__.py
@@ -0,0 +1,20 @@
+import sys
+from pathlib import Path
+
+import coverage
+from qgis.testing import unittest
+
+testfolder = Path(__file__).parent
+
+
+def run_all():
+ test_loader = unittest.defaultTestLoader
+ test_suite = test_loader.discover(".", pattern="test_*.py")
+
+ cov = coverage.Coverage(config_file=testfolder.parent / ".coveragerc")
+ cov.start()
+ unittest.TextTestRunner(verbosity=3, stream=sys.stdout).run(test_suite)
+
+ cov.stop()
+ cov.save()
+ cov.xml_report(outfile=testfolder / "coverage.xml")
diff --git a/ribasim_qgis/tests/ui/__init__.py b/ribasim_qgis/tests/ui/__init__.py
new file mode 100644
index 000000000..e69de29bb
diff --git a/ribasim_qgis/ui_tests/test_load_plugin.py b/ribasim_qgis/tests/ui/test_load_plugin.py
similarity index 100%
rename from ribasim_qgis/ui_tests/test_load_plugin.py
rename to ribasim_qgis/tests/ui/test_load_plugin.py
diff --git a/ribasim_qgis/ui_tests/__init__.py b/ribasim_qgis/ui_tests/__init__.py
deleted file mode 100644
index 81e3b194e..000000000
--- a/ribasim_qgis/ui_tests/__init__.py
+++ /dev/null
@@ -1,9 +0,0 @@
-import sys
-
-from qgis.testing import unittest
-
-
-def run_all():
- test_loader = unittest.defaultTestLoader
- test_suite = test_loader.discover(".", pattern="test_*.py")
- unittest.TextTestRunner(verbosity=3, stream=sys.stdout).run(test_suite)
From 23bbbbc1383b0f5f7b8e9c59c485124509bd5e4a Mon Sep 17 00:00:00 2001
From: Bart de Koning <74617371+SouthEndMusic@users.noreply.github.com>
Date: Thu, 29 Aug 2024 15:53:24 +0200
Subject: [PATCH 5/5] Re-enable `BackTracking` (#1761)
`BackTracking` as relaxation is now enabled again, with a thin wrapper
to reject it when the residual gets worse. Upstream issue:
https://github.com/SciML/OrdinaryDiffEq.jl/issues/2442
---
Manifest.toml | 12 ++----
core/Project.toml | 2 +
core/ext/RibasimMakieExt.jl | 34 ++++++---------
core/src/Ribasim.jl | 13 +++++-
core/src/config.jl | 6 ++-
core/src/model.jl | 4 +-
core/src/read.jl | 10 +++--
core/src/solve.jl | 11 +++++
core/src/util.jl | 82 +++++++++++++++++++++++++++++++++++--
core/test/main_test.jl | 1 -
10 files changed, 133 insertions(+), 42 deletions(-)
diff --git a/Manifest.toml b/Manifest.toml
index 2512ea2a4..1413e6984 100644
--- a/Manifest.toml
+++ b/Manifest.toml
@@ -2,7 +2,7 @@
julia_version = "1.10.4"
manifest_format = "2.0"
-project_hash = "c2cb085c326f61a96abd1a295e6fa775c585beba"
+project_hash = "a410a350a7b0c63bc6696029509aa68c14023275"
[[deps.ADTypes]]
git-tree-sha1 = "6778bcc27496dae5723ff37ee30af451db8b35fe"
@@ -1070,7 +1070,7 @@ uuid = "ca575930-c2e3-43a9-ace4-1e988b2c1908"
version = "1.2.0"
[[deps.NonlinearSolve]]
-deps = ["ADTypes", "ArrayInterface", "ConcreteStructs", "DiffEqBase", "FastBroadcast", "FastClosures", "FiniteDiff", "ForwardDiff", "LazyArrays", "LineSearches", "LinearAlgebra", "LinearSolve", "MaybeInplace", "PrecompileTools", "Preferences", "Printf", "RecursiveArrayTools", "Reexport", "SciMLBase", "SimpleNonlinearSolve", "SparseArrays", "SparseDiffTools", "StaticArraysCore", "SymbolicIndexingInterface", "TimerOutputs"]
+deps = ["ADTypes", "ArrayInterface", "ConcreteStructs", "DiffEqBase", "FastBroadcast", "FastClosures", "FiniteDiff", "ForwardDiff", "LazyArrays", "LineSearches", "LinearAlgebra", "LinearSolve", "MaybeInplace", "PrecompileTools", "Preferences", "Printf", "RecursiveArrayTools", "Reexport", "SciMLBase", "SimpleNonlinearSolve", "SparseArrays", "SparseDiffTools", "StaticArraysCore", "SymbolicIndexingInterface"]
git-tree-sha1 = "3adb1e5945b5a6b1eaee754077f25ccc402edd7f"
uuid = "8913a72c-1f9b-4ce2-8d82-65094dcecaec"
version = "3.13.1"
@@ -1302,7 +1302,7 @@ uuid = "295af30f-e4ad-537b-8983-00126c2a3abe"
version = "3.5.18"
[[deps.Ribasim]]
-deps = ["Accessors", "Arrow", "BasicModelInterface", "CodecZstd", "ComponentArrays", "Configurations", "DBInterface", "DataInterpolations", "DataStructures", "Dates", "DiffEqCallbacks", "EnumX", "FiniteDiff", "ForwardDiff", "Graphs", "HiGHS", "IterTools", "JuMP", "Legolas", "LinearSolve", "Logging", "LoggingExtras", "MetaGraphsNext", "OrdinaryDiffEq", "PreallocationTools", "SQLite", "SciMLBase", "SparseArrays", "SparseConnectivityTracer", "StructArrays", "Tables", "TerminalLoggers", "TranscodingStreams"]
+deps = ["Accessors", "Arrow", "BasicModelInterface", "CodecZstd", "ComponentArrays", "Configurations", "DBInterface", "DataInterpolations", "DataStructures", "Dates", "DiffEqCallbacks", "EnumX", "FiniteDiff", "ForwardDiff", "Graphs", "HiGHS", "IterTools", "JuMP", "Legolas", "LineSearches", "LinearSolve", "Logging", "LoggingExtras", "MetaGraphsNext", "OrdinaryDiffEq", "PreallocationTools", "SQLite", "SciMLBase", "SparseArrays", "SparseConnectivityTracer", "StructArrays", "Tables", "TerminalLoggers", "TranscodingStreams"]
path = "core"
uuid = "aac5e3d9-0b8f-4d4f-8241-b1a7a9632635"
version = "2024.10.0"
@@ -1669,12 +1669,6 @@ weakdeps = ["RecipesBase"]
[deps.TimeZones.extensions]
TimeZonesRecipesBaseExt = "RecipesBase"
-[[deps.TimerOutputs]]
-deps = ["ExprTools", "Printf"]
-git-tree-sha1 = "5a13ae8a41237cff5ecf34f73eb1b8f42fff6531"
-uuid = "a759f4b9-e2f1-59dc-863e-4aeb61b1ea8f"
-version = "0.5.24"
-
[[deps.TranscodingStreams]]
git-tree-sha1 = "d73336d81cafdc277ff45558bb7eaa2b04a8e472"
uuid = "3bb67fe8-82b1-5028-8e26-92a6c54297fa"
diff --git a/core/Project.toml b/core/Project.toml
index ce3e4882c..cf25cc035 100644
--- a/core/Project.toml
+++ b/core/Project.toml
@@ -24,6 +24,7 @@ HiGHS = "87dc4568-4c63-4d18-b0c0-bb2238e4078b"
IterTools = "c8e1da08-722c-5040-9ed9-7db0dc04731e"
JuMP = "4076af6c-e467-56ae-b986-b466b2749572"
Legolas = "741b9549-f6ed-4911-9fbf-4a1c0c97f0cd"
+LineSearches = "d3d80556-e9d4-5f37-9878-2ab0fcc64255"
LinearSolve = "7ed4a6bd-45f5-4d41-b270-4a48e9bafcae"
Logging = "56ddb016-857b-54e1-b83d-db4d58db5568"
LoggingExtras = "e6f89c97-d47a-5376-807f-9c37f3926c36"
@@ -70,6 +71,7 @@ IOCapture = "0.2"
IterTools = "1.4"
JuMP = "1.15"
Legolas = "0.5"
+LineSearches = "7"
LinearSolve = "2.24"
Logging = "<0.0.1, 1"
LoggingExtras = "1"
diff --git a/core/ext/RibasimMakieExt.jl b/core/ext/RibasimMakieExt.jl
index 97077bab8..7c21330ae 100644
--- a/core/ext/RibasimMakieExt.jl
+++ b/core/ext/RibasimMakieExt.jl
@@ -1,13 +1,13 @@
module RibasimMakieExt
using DataFrames: DataFrame
-using Makie: Figure, Axis, lines!, axislegend
+using Makie: Figure, Axis, scatterlines!, axislegend
using Ribasim: Ribasim, Model
function Ribasim.plot_basin_data!(model::Model, ax::Axis, column::Symbol)
basin_data = DataFrame(Ribasim.basin_table(model))
for node_id in unique(basin_data.node_id)
group = filter(:node_id => ==(node_id), basin_data)
- lines!(ax, group.time, getproperty(group, column); label = "Basin #$node_id")
+ scatterlines!(ax, group.time, getproperty(group, column); label = "Basin #$node_id")
end
axislegend(ax)
@@ -23,31 +23,23 @@ function Ribasim.plot_basin_data(model::Model)
f
end
-function Ribasim.plot_flow!(
- model::Model,
- ax::Axis,
- edge_id::Int32;
- skip_conservative_out = false,
-)
+function Ribasim.plot_flow!(model::Model, ax::Axis, edge_metadata::Ribasim.EdgeMetadata)
flow_data = DataFrame(Ribasim.flow_table(model))
- flow_data = filter(:edge_id => ==(edge_id), flow_data)
- first_row = first(flow_data)
- # Skip outflows of conservative nodes because these are the same as the inflows
- if skip_conservative_out &&
- Ribasim.NodeType.T(first_row.from_node_type) in Ribasim.conservative_nodetypes
- return nothing
- end
- label = "$(first_row.from_node_type) #$(first_row.from_node_id) → $(first_row.to_node_type) #$(first_row.to_node_id)"
- lines!(ax, flow_data.time, flow_data.flow_rate; label)
+ flow_data = filter(:edge_id => ==(edge_metadata.id), flow_data)
+ label = "$(edge_metadata.edge[1]) → $(edge_metadata.edge[2])"
+ scatterlines!(ax, flow_data.time, flow_data.flow_rate; label)
return nothing
end
-function Ribasim.plot_flow(model::Model)
+function Ribasim.plot_flow(model::Model; skip_conservative_out = true)
f = Figure()
ax = Axis(f[1, 1]; xlabel = "time", ylabel = "flow rate [m³s⁻¹]")
- edge_ids = unique(Ribasim.flow_table(model).edge_id)
- for edge_id in edge_ids
- Ribasim.plot_flow!(model, ax, edge_id; skip_conservative_out = true)
+ for edge_metadata in values(model.integrator.p.graph.edge_data)
+ if skip_conservative_out &&
+ edge_metadata.edge[1].type in Ribasim.conservative_nodetypes
+ continue
+ end
+ Ribasim.plot_flow!(model, ax, edge_metadata)
end
axislegend(ax)
f
diff --git a/core/src/Ribasim.jl b/core/src/Ribasim.jl
index 059a62d4e..01d735dd2 100644
--- a/core/src/Ribasim.jl
+++ b/core/src/Ribasim.jl
@@ -15,7 +15,15 @@ For more granular access, see:
module Ribasim
# Algorithms for solving ODEs.
-using OrdinaryDiffEq: OrdinaryDiffEq, OrdinaryDiffEqRosenbrockAdaptiveAlgorithm, get_du
+using OrdinaryDiffEq:
+ OrdinaryDiffEq,
+ OrdinaryDiffEqRosenbrockAdaptiveAlgorithm,
+ get_du,
+ AbstractNLSolver,
+ relax!,
+ _compute_rhs!,
+ calculate_residuals!
+using LineSearches: BackTracking
# Interface for defining and solving the ODE problem of the physical layer.
using SciMLBase:
@@ -31,7 +39,8 @@ using SciMLBase:
ODEProblem,
ODESolution,
VectorContinuousCallback,
- get_proposed_dt
+ get_proposed_dt,
+ DEIntegrator
# Automatically detecting the sparsity pattern of the Jacobian of water_balance!
# through operator overloading
diff --git a/core/src/config.jl b/core/src/config.jl
index 794a17477..bb024cd0f 100644
--- a/core/src/config.jl
+++ b/core/src/config.jl
@@ -230,7 +230,7 @@ const algorithms = Dict{String, Type}(
)
"Create an OrdinaryDiffEqAlgorithm from solver config"
-function algorithm(solver::Solver)::OrdinaryDiffEqAlgorithm
+function algorithm(solver::Solver; u0 = [])::OrdinaryDiffEqAlgorithm
algotype = get(algorithms, solver.algorithm, nothing)
if algotype === nothing
options = join(keys(algorithms), ", ")
@@ -239,7 +239,9 @@ function algorithm(solver::Solver)::OrdinaryDiffEqAlgorithm
end
kwargs = Dict{Symbol, Any}()
if algotype <: OrdinaryDiffEqNewtonAdaptiveAlgorithm
- kwargs[:nlsolve] = NLNewton(; relax = 0.1)
+ kwargs[:nlsolve] = NLNewton(;
+ relax = Ribasim.MonitoredBackTracking(; z_tmp = copy(u0), dz_tmp = copy(u0)),
+ )
end
# not all algorithms support this keyword
kwargs[:autodiff] = solver.autodiff
diff --git a/core/src/model.jl b/core/src/model.jl
index 13d898336..9c6a447be 100644
--- a/core/src/model.jl
+++ b/core/src/model.jl
@@ -37,7 +37,6 @@ function Model(config_path::AbstractString)::Model
end
function Model(config::Config)::Model
- alg = algorithm(config.solver)
db_path = input_path(config, config.database)
if !isfile(db_path)
@error "Database file not found" db_path
@@ -109,6 +108,9 @@ function Model(config::Config)::Model
u0 = ComponentVector{Float64}(; storage, integral)
du0 = zero(u0)
+ # The Solver algorithm
+ alg = algorithm(config.solver; u0)
+
# Synchronize level with storage
set_current_basin_properties!(parameters.basin, u0, du0)
diff --git a/core/src/read.jl b/core/src/read.jl
index e76793e28..05102d64e 100644
--- a/core/src/read.jl
+++ b/core/src/read.jl
@@ -572,7 +572,8 @@ function Basin(db::DB, config::Config, graph::MetaGraph)::Basin
error("Invalid Basin / profile table.")
end
- level_to_area = LinearInterpolation.(area, level; extrapolate = true)
+ level_to_area =
+ LinearInterpolation.(area, level; extrapolate = true, cache_parameters = true)
storage_to_level = invert_integral.(level_to_area)
t_end = seconds_since(config.endtime, config.starttime)
@@ -921,6 +922,7 @@ function user_demand_static!(
fill(first_row.return_factor, 2),
return_factor_old.t;
extrapolate = true,
+ cache_parameters = true,
)
min_level[user_demand_idx] = first_row.min_level
@@ -1026,8 +1028,10 @@ function UserDemand(db::DB, config::Config, graph::MetaGraph)::UserDemand
]
demand_from_timeseries = fill(false, n_user)
allocated = fill(Inf, n_user, n_priority)
- return_factor =
- [LinearInterpolation(zeros(2), trivial_timespan) for i in eachindex(node_ids)]
+ return_factor = [
+ LinearInterpolation(zeros(2), trivial_timespan; cache_parameters = true) for
+ i in eachindex(node_ids)
+ ]
min_level = zeros(n_user)
# Process static table
diff --git a/core/src/solve.jl b/core/src/solve.jl
index cabfcd6e0..04aea58a7 100644
--- a/core/src/solve.jl
+++ b/core/src/solve.jl
@@ -51,9 +51,20 @@ function water_balance!(
# Formulate du (controlled by PidControl)
formulate_du_pid_controlled!(du, graph, pid_control)
+ # https://github.com/Deltares/Ribasim/issues/1705#issuecomment-2283293974
+ stop_declining_negative_storage!(du, u)
+
return nothing
end
+function stop_declining_negative_storage!(du, u)
+ for (i, s) in enumerate(u.storage)
+ if s < 0
+ du.storage[i] = max(du.storage[i], 0.0)
+ end
+ end
+end
+
function formulate_continuous_control!(du, p, t)::Nothing
(; compound_variable, target_ref, func) = p.continuous_control
diff --git a/core/src/util.jl b/core/src/util.jl
index 695f62435..90d197cf3 100644
--- a/core/src/util.jl
+++ b/core/src/util.jl
@@ -53,9 +53,18 @@ end
Compute the area and level of a basin given its storage.
"""
function get_area_and_level(basin::Basin, state_idx::Int, storage::T)::Tuple{T, T} where {T}
- level = basin.storage_to_level[state_idx](max(storage, 0.0))
- area = basin.level_to_area[state_idx](level)
-
+ storage_to_level = basin.storage_to_level[state_idx]
+ level_to_area = basin.level_to_area[state_idx]
+ if storage >= 0
+ level = storage_to_level(storage)
+ else
+ # Negative storage is not feasible and this yields a level
+ # below the basin bottom, but this does yield usable gradients
+ # for the non-linear solver
+ bottom = first(level_to_area.t)
+ level = bottom + derivative(storage_to_level, 0.0) * storage
+ end
+ area = level_to_area(level)
return area, level
end
@@ -887,3 +896,70 @@ end
(A::AbstractInterpolation)(t::GradientTracer) = t
reduction_factor(x::GradientTracer, threshold::Real) = x
relaxed_root(x::GradientTracer, threshold::Real) = x
+get_area_and_level(basin::Basin, state_idx::Int, storage::GradientTracer) = storage, storage
+stop_declining_negative_storage!(du, u::ComponentVector{<:GradientTracer}) = nothing
+
+@kwdef struct MonitoredBackTracking{B, V}
+ linesearch::B = BackTracking()
+ dz_tmp::V = []
+ z_tmp::V = []
+end
+
+"""
+Compute the residual of the non-linear solver, i.e. a measure of the
+error in the solution to the implicit equation defined by the solver algorithm
+"""
+function residual(z, integrator, nlsolver, f)
+ (; uprev, t, p, dt, opts, isdae) = integrator
+ (; tmp, ztmp, γ, α, cache, method) = nlsolver
+ (; ustep, atmp, tstep, k, invγdt, tstep, k, invγdt) = cache
+ if isdae
+ _uprev = get_dae_uprev(integrator, uprev)
+ b, ustep2 =
+ _compute_rhs!(tmp, ztmp, ustep, α, tstep, k, invγdt, p, _uprev, f::TF, z)
+ else
+ b, ustep2 =
+ _compute_rhs!(tmp, ztmp, ustep, γ, α, tstep, k, invγdt, method, p, dt, f, z)
+ end
+ calculate_residuals!(
+ atmp,
+ b,
+ uprev,
+ ustep2,
+ opts.abstol,
+ opts.reltol,
+ opts.internalnorm,
+ t,
+ )
+ ndz = opts.internalnorm(atmp, t)
+ return ndz
+end
+
+"""
+MonitoredBackTracing is a thin wrapper of BackTracking, making sure that
+the BackTracking relaxation is rejected if it results in a residual increase
+"""
+function OrdinaryDiffEq.relax!(
+ dz,
+ nlsolver::AbstractNLSolver,
+ integrator::DEIntegrator,
+ f,
+ linesearch::MonitoredBackTracking,
+)
+ (; linesearch, dz_tmp, z_tmp) = linesearch
+
+ # Store step before relaxation
+ @. dz_tmp = dz
+
+ # Apply relaxation and measure the residual change
+ @. z_tmp = nlsolver.z + dz
+ resid_before = residual(z_tmp, integrator, nlsolver, f)
+ relax!(dz, nlsolver, integrator, f, linesearch)
+ @. z_tmp = nlsolver.z + dz
+ resid_after = residual(z_tmp, integrator, nlsolver, f)
+
+ # If the residual increased due to the relaxation, reject it
+ if resid_after > resid_before
+ @. dz = dz_tmp
+ end
+end
diff --git a/core/test/main_test.jl b/core/test/main_test.jl
index 788e9c7c3..95dfac40e 100644
--- a/core/test/main_test.jl
+++ b/core/test/main_test.jl
@@ -24,7 +24,6 @@
@show backtrace
end
@test occursin("version in the TOML config file does not match", output)
- @test occursin("Info: Convergence bottlenecks in descending order of severity:", output)
end
@testitem "main error logging" begin
+
+
# Inspecting results {#sec-results}
Before inspecting the results, verify that the run was successfully and the output files are there.
diff --git a/python/ribasim/ribasim/styles/EdgeStyle.qml b/python/ribasim/ribasim/styles/EdgeStyle.qml
index acbb8381b..5b9bee800 100644
--- a/python/ribasim/ribasim/styles/EdgeStyle.qml
+++ b/python/ribasim/ribasim/styles/EdgeStyle.qml
@@ -555,7 +555,7 @@
-
+
diff --git a/ribasim_qgis/core/nodes.py b/ribasim_qgis/core/nodes.py
index f257fe8ee..38fbbeee7 100644
--- a/ribasim_qgis/core/nodes.py
+++ b/ribasim_qgis/core/nodes.py
@@ -123,6 +123,15 @@ def set_dropdown(self, name: str, options: set[str]) -> None:
)
layer.setEditorWidgetSetup(index, setup)
+ def set_unique(self, name: str) -> None:
+ layer = self.layer
+ index = layer.fields().indexFromName(name)
+ setup = QgsEditorWidgetSetup(
+ "UniqueValues",
+ {},
+ )
+ layer.setEditorWidgetSetup(index, setup)
+
def set_read_only(self) -> None:
pass
@@ -134,8 +143,11 @@ def layer_from_geopackage(self) -> QgsVectorLayer:
self.layer = QgsVectorLayer(
f"{self._path}|layername={self.input_type()}", self.input_type()
)
- # Load style from database if exists
- self.layer.loadDefaultStyle()
+ # Load style from database if exists, otherwise load and save default qml style
+ _, success = self.layer.loadDefaultStyle()
+ if not success:
+ self.load_default_style()
+ self.save_style()
# Connect signal to save style to database when changed
self.layer.styleChanged.connect(self.save_style)
return self.layer
@@ -214,6 +226,7 @@ def set_editor_widget(self) -> None:
layer = self.layer
node_type_field_index = layer.fields().indexFromName("node_type")
self.set_dropdown("node_type", NONSPATIALNODETYPES)
+ self.set_unique("node_id")
layer_form_config = layer.editFormConfig()
layer_form_config.setReuseLastValue(node_type_field_index, True)
@@ -267,6 +280,7 @@ def set_editor_widget(self) -> None:
layer = self.layer
self.set_dropdown("edge_type", EDGETYPES)
+ self.set_unique("edge_id")
layer_form_config = layer.editFormConfig()
layer.setEditFormConfig(layer_form_config)
@@ -859,7 +873,7 @@ def attributes(cls) -> list[QgsField]:
}
NONSPATIALNODETYPES: set[str] = {
cls.nodetype() for cls in Input.__subclasses__() if not cls.is_spatial()
-}
+} | {"Terminal"}
EDGETYPES = {"flow", "control"}
SPATIALCONTROLNODETYPES = {
"ContinuousControl",
diff --git a/ribasim_qgis/widgets/dataset_widget.py b/ribasim_qgis/widgets/dataset_widget.py
index d43eff23a..b84656e16 100644
--- a/ribasim_qgis/widgets/dataset_widget.py
+++ b/ribasim_qgis/widgets/dataset_widget.py
@@ -8,6 +8,7 @@
from __future__ import annotations
from datetime import datetime
+from functools import partial
from pathlib import Path
from typing import Any, cast
@@ -27,8 +28,11 @@
QWidget,
)
from qgis.core import (
+ QgsEditorWidgetSetup,
+ QgsFeatureRequest,
QgsMapLayer,
QgsProject,
+ QgsRelation,
QgsVectorLayer,
)
@@ -208,6 +212,32 @@ def add_selection_to_qgis(self) -> None:
for item in selection:
self.add_item_to_qgis(item)
+ @staticmethod
+ def add_relationship(from_layer, to_layer_id, name, fk="node_id") -> None:
+ rel = QgsRelation()
+ rel.setReferencingLayer(from_layer.id())
+ rel.setReferencedLayer(to_layer_id)
+ rel.setName(name)
+ rel.setStrength(rel.RelationStrength.Composition) # type: ignore
+ rel.addFieldPair(fk, "node_id")
+ rel.generateId()
+ instance = QgsProject.instance()
+ assert instance is not None
+ rel_manager = instance.relationManager()
+ assert rel_manager is not None
+ rel_manager.addRelation(rel)
+
+ # Also use the relationship as an editor widget
+ field_index = from_layer.fields().indexFromName(fk)
+ setup = QgsEditorWidgetSetup(
+ "RelationReference",
+ {
+ "Relation": rel.id(),
+ "MapIdentification": True,
+ },
+ )
+ from_layer.setEditorWidgetSetup(field_index, setup)
+
def load_geopackage(self) -> None:
"""Load the layers of a GeoPackage into the Layers Panel"""
self.dataset_tree.clear()
@@ -221,25 +251,66 @@ def load_geopackage(self) -> None:
node = nodes.pop("Node")
item = self.dataset_tree.add_node_layer(node)
self.add_item_to_qgis(item)
+ # Make sure node_id shows up in relationships
+ node.layer.setDisplayExpression("node_id")
edge = nodes.pop("Edge")
item = self.dataset_tree.add_node_layer(edge)
self.add_item_to_qgis(item)
+ self.add_relationship(
+ edge.layer, node.layer.id(), "EdgeFromNode", "from_node_id"
+ )
+ self.add_relationship(edge.layer, node.layer.id(), "EdgeToNode", "to_node_id")
basin_area_layer = nodes.pop("Basin / area", None)
if basin_area_layer is not None:
item = self.dataset_tree.add_node_layer(basin_area_layer)
self.add_item_to_qgis(item)
+ self.add_relationship(
+ basin_area_layer.layer, node.layer.id(), "Basin / area"
+ )
# Add the remaining layers
- for node_layer in nodes.values():
+ for table_name, node_layer in nodes.items():
item = self.dataset_tree.add_node_layer(node_layer)
self.add_item_to_qgis(item)
+ self.add_relationship(node_layer.layer, node.layer.id(), table_name)
# Connect node and edge layer to derive connectivities.
self.node_layer = node.layer
+ assert self.node_layer is not None
self.edge_layer = edge.layer
self.edge_layer.editingStopped.connect(self.connect_nodes)
+
+ def filterbyrel(relationships, feature_ids):
+ """Filter all related tables by the selected features in the node table."""
+ ids = []
+ selection = QgsFeatureRequest().setFilterFids(feature_ids)
+ for rel in relationships:
+ for feature in rel.referencedLayer().getFeatures(selection):
+ ids.extend(f.id() for f in rel.getRelatedFeatures(feature))
+
+ rel.referencingLayer().selectByIds(ids)
+
+ # When the Node selection changes, filter all related tables
+ edge_rels = []
+ instance = QgsProject.instance()
+ assert instance is not None
+ rel_manager = instance.relationManager()
+ assert rel_manager is not None
+ for rel in rel_manager.relations().values():
+ # Edge relations are special, they have two references to the Node table
+ referencing = rel.referencingLayer()
+ referenced = rel.referencedLayer()
+ assert referencing is not None
+ assert referenced is not None
+
+ if referencing.name() == "Edge":
+ edge_rels.append(rel)
+ else:
+ referenced.selectionChanged.connect(partial(filterbyrel, [rel]))
+
+ self.node_layer.selectionChanged.connect(partial(filterbyrel, edge_rels))
return
def new_model(self) -> None:
diff --git a/ribasim_qgis/widgets/nodes_widget.py b/ribasim_qgis/widgets/nodes_widget.py
index 689e0e78a..cdaa293ae 100644
--- a/ribasim_qgis/widgets/nodes_widget.py
+++ b/ribasim_qgis/widgets/nodes_widget.py
@@ -67,5 +67,7 @@ def new_node_layer(self, node_type: str) -> None:
node.write()
# Add to QGIS
self.ribasim_widget.add_layer(node.layer, "Ribasim Input", labels=node.labels)
+ # Setup relationship
+ self.ribasim_widget.add_relationship(node.layer, node_type)
# Add to dataset tree
self.ribasim_widget.add_node_layer(node)
diff --git a/ribasim_qgis/widgets/ribasim_widget.py b/ribasim_qgis/widgets/ribasim_widget.py
index 2635865e8..8af4689c4 100644
--- a/ribasim_qgis/widgets/ribasim_widget.py
+++ b/ribasim_qgis/widgets/ribasim_widget.py
@@ -106,7 +106,7 @@ def create_groups(self, name: str) -> None:
assert project is not None
root = project.layerTreeRoot()
assert root is not None
- self.group = root.addGroup(name)
+ self.group = root.insertGroup(0, name) # insert at the top
self.create_subgroup(name, "Ribasim Input")
def add_to_group(self, maplayer: Any, destination: str, on_top: bool):
@@ -182,3 +182,7 @@ def add_layer(
self.add_to_group(maplayer, destination, on_top)
return maplayer
+
+ def add_relationship(self, new_layer, name: str) -> None:
+ assert self.node_layer is not None
+ self.__dataset_widget.add_relationship(new_layer, self.node_layer.id(), name)
From 4cf93b5ec7e38135f4e870d0bb714d0a06890068 Mon Sep 17 00:00:00 2001
From: Bart de Koning <74617371+SouthEndMusic@users.noreply.github.com>
Date: Tue, 27 Aug 2024 18:43:58 +0200
Subject: [PATCH 2/5] Give no bottleneck warning if no bottleneck basins were
found (#1770)
Fixes https://github.com/Deltares/Ribasim/issues/1759
---
core/src/logging.jl | 4 +++-
1 file changed, 3 insertions(+), 1 deletion(-)
diff --git a/core/src/logging.jl b/core/src/logging.jl
index 18f474de9..13f3dc020 100644
--- a/core/src/logging.jl
+++ b/core/src/logging.jl
@@ -46,7 +46,9 @@ function log_bottlenecks(model; converged::Bool)
end
push!(errors, node_id => error)
end
- @logmsg level "Convergence bottlenecks in descending order of severity:" errors...
+ if !isempty(errors)
+ @logmsg level "Convergence bottlenecks in descending order of severity:" errors...
+ end
else
algorithm = model.config.solver.algorithm
@logmsg level "Convergence bottlenecks are not shown for the chosen solver algorithm." algorithm
From 7ec16c76f0ea3931e703cee26cd4b80e92fc0152 Mon Sep 17 00:00:00 2001
From: Maarten Pronk
Date: Thu, 29 Aug 2024 13:43:48 +0200
Subject: [PATCH 3/5] Error when the toml or database can't be found. (#1773)
Fixes #1652
---
python/ribasim/ribasim/model.py | 9 ++++++++-
python/ribasim/tests/test_io.py | 1 +
python/ribasim/tests/test_model.py | 17 +++++++++++++++++
3 files changed, 26 insertions(+), 1 deletion(-)
diff --git a/python/ribasim/ribasim/model.py b/python/ribasim/ribasim/model.py
index 11c50da96..947738e80 100644
--- a/python/ribasim/ribasim/model.py
+++ b/python/ribasim/ribasim/model.py
@@ -253,6 +253,8 @@ def read(cls, filepath: str | PathLike[str]) -> "Model":
filepath : str | PathLike[str]
The path to the TOML file.
"""
+ if not Path(filepath).is_file():
+ raise FileNotFoundError(f"File '{filepath}' does not exist.")
return cls(filepath=filepath) # type: ignore
def write(self, filepath: str | PathLike[str]) -> Path:
@@ -290,7 +292,12 @@ def _load(cls, filepath: Path | None) -> dict[str, Any]:
directory = filepath.parent / config.get("input_dir", ".")
context_file_loading.get()["directory"] = directory
- context_file_loading.get()["database"] = directory / "database.gpkg"
+ db_path = directory / "database.gpkg"
+
+ if not db_path.is_file():
+ raise FileNotFoundError(f"Database file '{db_path}' does not exist.")
+
+ context_file_loading.get()["database"] = db_path
return config
else:
diff --git a/python/ribasim/tests/test_io.py b/python/ribasim/tests/test_io.py
index 60d977316..5aedd70da 100644
--- a/python/ribasim/tests/test_io.py
+++ b/python/ribasim/tests/test_io.py
@@ -299,5 +299,6 @@ def test_datetime_timezone():
def test_minimal_toml():
# Check if the TOML used in QGIS tests is still valid.
toml_path = Path(__file__).parents[3] / "ribasim_qgis/tests/data/simple_valid.toml"
+ (toml_path.parent / "database.gpkg").touch() # database file must exist for `read`
model = ribasim.Model.read(toml_path)
assert model.crs == "EPSG:28992"
diff --git a/python/ribasim/tests/test_model.py b/python/ribasim/tests/test_model.py
index 6b96988c9..7f2dfdee5 100644
--- a/python/ribasim/tests/test_model.py
+++ b/python/ribasim/tests/test_model.py
@@ -4,6 +4,7 @@
import numpy as np
import pandas as pd
import pytest
+import tomli_w
import xugrid
from pydantic import ValidationError
from pyproj import CRS
@@ -228,3 +229,19 @@ def test_styles(tabulated_rating_curve: Model, tmp_path):
model.write(tmp_path / "basic" / "ribasim.toml")
with connect(tmp_path / "basic" / "database.gpkg") as conn:
assert conn.execute("SELECT COUNT(*) FROM layer_styles").fetchone()[0] == 3
+
+
+def test_non_existent_files(tmp_path):
+ with pytest.raises(
+ FileNotFoundError, match="File 'non_existent_file.toml' does not exist."
+ ):
+ Model.read("non_existent_file.toml")
+
+ # Create a TOML file without a database.gpkg
+ content = {"input_path": str(tmp_path)}
+ toml_path = tmp_path / "test.toml"
+ with open(toml_path, "wb") as f:
+ tomli_w.dump(content, f)
+
+ with pytest.raises(FileNotFoundError, match=r"Database file .* does not exist\."):
+ Model.read(toml_path)
From b80a79addac96c8ad1c7f4657af871da3c69d627 Mon Sep 17 00:00:00 2001
From: Maarten Pronk
Date: Thu, 29 Aug 2024 13:46:53 +0200
Subject: [PATCH 4/5] Enable Docker for QGIS tests (once more) (#1774)
Fixes #1751
Changes since last time:
- Also includes coverage
- Also runs other QGIS tests
---
.docker/compose.yml | 12 ++++++
.docker/start.sh | 7 ++++
.docker/stop.sh | 6 +++
.docker/test.sh | 4 ++
.github/workflows/qgis.yml | 40 ++++++++-----------
pixi.toml | 5 ++-
ribasim_qgis/.coveragerc | 8 ++--
ribasim_qgis/scripts/run_qgis_ui_tests.py | 2 +-
ribasim_qgis/tests/__init__.py | 20 ++++++++++
ribasim_qgis/tests/ui/__init__.py | 0
.../ui}/test_load_plugin.py | 0
ribasim_qgis/ui_tests/__init__.py | 9 -----
12 files changed, 74 insertions(+), 39 deletions(-)
create mode 100644 .docker/compose.yml
create mode 100755 .docker/start.sh
create mode 100755 .docker/stop.sh
create mode 100755 .docker/test.sh
create mode 100644 ribasim_qgis/tests/__init__.py
create mode 100644 ribasim_qgis/tests/ui/__init__.py
rename ribasim_qgis/{ui_tests => tests/ui}/test_load_plugin.py (100%)
delete mode 100644 ribasim_qgis/ui_tests/__init__.py
diff --git a/.docker/compose.yml b/.docker/compose.yml
new file mode 100644
index 000000000..8f6f42924
--- /dev/null
+++ b/.docker/compose.yml
@@ -0,0 +1,12 @@
+version: '2'
+
+services:
+ qgis:
+ image: qgis/qgis:release-3_34
+ container_name: qgis
+ volumes:
+ - ../ribasim_qgis/:/tests_directory/ribasim_qgis
+ environment:
+ - CI=true
+ - DISPLAY=:99
+ tty: true
diff --git a/.docker/start.sh b/.docker/start.sh
new file mode 100755
index 000000000..6c583b16b
--- /dev/null
+++ b/.docker/start.sh
@@ -0,0 +1,7 @@
+#!/usr/bin/env bash
+set -eux
+
+docker compose -f compose.yml up -d --force-recreate --remove-orphans
+echo "Installation of the plugin Ribasim"
+docker exec -t qgis sh -c "qgis_setup.sh ribasim_qgis"
+echo "Containers are running"
diff --git a/.docker/stop.sh b/.docker/stop.sh
new file mode 100755
index 000000000..08afc0ec6
--- /dev/null
+++ b/.docker/stop.sh
@@ -0,0 +1,6 @@
+#!/usr/bin/env bash
+set -eux
+
+echo 'Stopping/killing containers'
+docker compose -f compose.yml kill
+docker compose -f compose.yml rm -f
diff --git a/.docker/test.sh b/.docker/test.sh
new file mode 100755
index 000000000..b1ce0ec3b
--- /dev/null
+++ b/.docker/test.sh
@@ -0,0 +1,4 @@
+#!/usr/bin/env bash
+set -eux
+
+docker exec -t qgis sh -c "cd /tests_directory && xvfb-run -a qgis_testrunner.sh ribasim_qgis.tests"
diff --git a/.github/workflows/qgis.yml b/.github/workflows/qgis.yml
index fc9b302de..3275c8db4 100644
--- a/.github/workflows/qgis.yml
+++ b/.github/workflows/qgis.yml
@@ -8,27 +8,21 @@ on:
pull_request:
paths-ignore: [".teamcity/**"]
merge_group:
-concurrency:
- group: ${{ github.workflow }}-${{ github.ref }}
- cancel-in-progress: true
jobs:
- test:
- name: QGIS plugin ${{ matrix.os }}
- runs-on: ${{ matrix.os }}
- strategy:
- matrix:
- os:
- - ubuntu-latest
- - macOS-latest
- - windows-latest
- steps:
- - uses: actions/checkout@v4
- - uses: prefix-dev/setup-pixi@v0.8.1
- with:
- pixi-version: "latest"
- - name: Run tests
- run: pixi run test-ribasim-qgis-cov
- - name: Upload coverage to Codecov
- uses: codecov/codecov-action@v4
- with:
- token: ${{ secrets.CODECOV_TOKEN }}
+ test-qgis:
+ name: "Test"
+ runs-on: ubuntu-latest
+ defaults:
+ run:
+ working-directory: .docker
+ steps:
+ - uses: actions/checkout@v4
+ - uses: prefix-dev/setup-pixi@v0.8.1
+ with:
+ pixi-version: "latest"
+ - name: Run tests
+ run: pixi run test-ribasim-qgis-docker
+ - name: Upload coverage to Codecov
+ uses: codecov/codecov-action@v4
+ with:
+ token: ${{ secrets.CODECOV_TOKEN }}
diff --git a/pixi.toml b/pixi.toml
index e16056b63..346715472 100644
--- a/pixi.toml
+++ b/pixi.toml
@@ -128,6 +128,7 @@ install-ribasim-qgis = "python ribasim_qgis/scripts/install_ribasim_qgis.py"
install-imod-qgis = "python ribasim_qgis/scripts/install_qgis_plugin.py iMOD && python ribasim_qgis/scripts/enable_plugin.py imodqgis"
install-plugin-reloader-qgis = "python ribasim_qgis/scripts/install_qgis_plugin.py \"Plugin Reloader\" && python ribasim_qgis/scripts/enable_plugin.py plugin_reloader"
install-debugvs-qgis = "python ribasim_qgis/scripts/install_qgis_plugin.py debugvs==0.7 && python ribasim_qgis/scripts/enable_plugin.py debug_vs"
+test-ribasim-qgis-docker = { cmd = "sh ./start.sh; sh ./test.sh; sh ./stop.sh", cwd = ".docker" }
install-qgis-plugins = { depends_on = [
"install-plugin-reloader-qgis",
"install-ribasim-qgis",
@@ -137,10 +138,10 @@ install-qgis-plugins = { depends_on = [
test-ribasim-qgis-ui = { cmd = "python ribasim_qgis/scripts/run_qgis_ui_tests.py", depends_on = [
"install-ribasim-qgis",
] }
-test-ribasim-qgis = { cmd = "pytest --numprocesses=auto ribasim_qgis/tests", depends_on = [
+test-ribasim-qgis = { cmd = "pytest --numprocesses=auto ribasim_qgis/tests/core", depends_on = [
"install-ribasim-qgis",
] }
-test-ribasim-qgis-cov = { cmd = "pytest --numprocesses=auto --cov=ribasim_qgis --cov-report=xml --cov-config=ribasim_qgis/.coveragerc ribasim_qgis/tests", depends_on = [
+test-ribasim-qgis-cov = { cmd = "pytest --numprocesses=auto --cov=ribasim_qgis --cov-report=xml --cov-config=ribasim_qgis/.coveragerc ribasim_qgis/tests/core", depends_on = [
"install-ribasim-qgis",
] }
mypy-ribasim-qgis = "mypy ribasim_qgis"
diff --git a/ribasim_qgis/.coveragerc b/ribasim_qgis/.coveragerc
index 011bc9fa5..e14c37159 100644
--- a/ribasim_qgis/.coveragerc
+++ b/ribasim_qgis/.coveragerc
@@ -1,6 +1,6 @@
[run]
+source = .
omit =
- ribasim_qgis/resources.py
- ribasim_qgis/tests/*
- ribasim_qgis/tomllib/*
- ribasim_qgis/ui_tests/*
+ */resources.py
+ */tests/*
+ */tomllib/*
diff --git a/ribasim_qgis/scripts/run_qgis_ui_tests.py b/ribasim_qgis/scripts/run_qgis_ui_tests.py
index bee8e1096..04fda7c4d 100644
--- a/ribasim_qgis/scripts/run_qgis_ui_tests.py
+++ b/ribasim_qgis/scripts/run_qgis_ui_tests.py
@@ -9,7 +9,7 @@
"--nologo",
"--code",
"ribasim_qgis/scripts/qgis_testrunner.py",
- "ribasim_qgis.ui_tests",
+ "ribasim_qgis.tests",
],
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
diff --git a/ribasim_qgis/tests/__init__.py b/ribasim_qgis/tests/__init__.py
new file mode 100644
index 000000000..7034cc660
--- /dev/null
+++ b/ribasim_qgis/tests/__init__.py
@@ -0,0 +1,20 @@
+import sys
+from pathlib import Path
+
+import coverage
+from qgis.testing import unittest
+
+testfolder = Path(__file__).parent
+
+
+def run_all():
+ test_loader = unittest.defaultTestLoader
+ test_suite = test_loader.discover(".", pattern="test_*.py")
+
+ cov = coverage.Coverage(config_file=testfolder.parent / ".coveragerc")
+ cov.start()
+ unittest.TextTestRunner(verbosity=3, stream=sys.stdout).run(test_suite)
+
+ cov.stop()
+ cov.save()
+ cov.xml_report(outfile=testfolder / "coverage.xml")
diff --git a/ribasim_qgis/tests/ui/__init__.py b/ribasim_qgis/tests/ui/__init__.py
new file mode 100644
index 000000000..e69de29bb
diff --git a/ribasim_qgis/ui_tests/test_load_plugin.py b/ribasim_qgis/tests/ui/test_load_plugin.py
similarity index 100%
rename from ribasim_qgis/ui_tests/test_load_plugin.py
rename to ribasim_qgis/tests/ui/test_load_plugin.py
diff --git a/ribasim_qgis/ui_tests/__init__.py b/ribasim_qgis/ui_tests/__init__.py
deleted file mode 100644
index 81e3b194e..000000000
--- a/ribasim_qgis/ui_tests/__init__.py
+++ /dev/null
@@ -1,9 +0,0 @@
-import sys
-
-from qgis.testing import unittest
-
-
-def run_all():
- test_loader = unittest.defaultTestLoader
- test_suite = test_loader.discover(".", pattern="test_*.py")
- unittest.TextTestRunner(verbosity=3, stream=sys.stdout).run(test_suite)
From 23bbbbc1383b0f5f7b8e9c59c485124509bd5e4a Mon Sep 17 00:00:00 2001
From: Bart de Koning <74617371+SouthEndMusic@users.noreply.github.com>
Date: Thu, 29 Aug 2024 15:53:24 +0200
Subject: [PATCH 5/5] Re-enable `BackTracking` (#1761)
`BackTracking` as relaxation is now enabled again, with a thin wrapper
to reject it when the residual gets worse. Upstream issue:
https://github.com/SciML/OrdinaryDiffEq.jl/issues/2442
---
Manifest.toml | 12 ++----
core/Project.toml | 2 +
core/ext/RibasimMakieExt.jl | 34 ++++++---------
core/src/Ribasim.jl | 13 +++++-
core/src/config.jl | 6 ++-
core/src/model.jl | 4 +-
core/src/read.jl | 10 +++--
core/src/solve.jl | 11 +++++
core/src/util.jl | 82 +++++++++++++++++++++++++++++++++++--
core/test/main_test.jl | 1 -
10 files changed, 133 insertions(+), 42 deletions(-)
diff --git a/Manifest.toml b/Manifest.toml
index 2512ea2a4..1413e6984 100644
--- a/Manifest.toml
+++ b/Manifest.toml
@@ -2,7 +2,7 @@
julia_version = "1.10.4"
manifest_format = "2.0"
-project_hash = "c2cb085c326f61a96abd1a295e6fa775c585beba"
+project_hash = "a410a350a7b0c63bc6696029509aa68c14023275"
[[deps.ADTypes]]
git-tree-sha1 = "6778bcc27496dae5723ff37ee30af451db8b35fe"
@@ -1070,7 +1070,7 @@ uuid = "ca575930-c2e3-43a9-ace4-1e988b2c1908"
version = "1.2.0"
[[deps.NonlinearSolve]]
-deps = ["ADTypes", "ArrayInterface", "ConcreteStructs", "DiffEqBase", "FastBroadcast", "FastClosures", "FiniteDiff", "ForwardDiff", "LazyArrays", "LineSearches", "LinearAlgebra", "LinearSolve", "MaybeInplace", "PrecompileTools", "Preferences", "Printf", "RecursiveArrayTools", "Reexport", "SciMLBase", "SimpleNonlinearSolve", "SparseArrays", "SparseDiffTools", "StaticArraysCore", "SymbolicIndexingInterface", "TimerOutputs"]
+deps = ["ADTypes", "ArrayInterface", "ConcreteStructs", "DiffEqBase", "FastBroadcast", "FastClosures", "FiniteDiff", "ForwardDiff", "LazyArrays", "LineSearches", "LinearAlgebra", "LinearSolve", "MaybeInplace", "PrecompileTools", "Preferences", "Printf", "RecursiveArrayTools", "Reexport", "SciMLBase", "SimpleNonlinearSolve", "SparseArrays", "SparseDiffTools", "StaticArraysCore", "SymbolicIndexingInterface"]
git-tree-sha1 = "3adb1e5945b5a6b1eaee754077f25ccc402edd7f"
uuid = "8913a72c-1f9b-4ce2-8d82-65094dcecaec"
version = "3.13.1"
@@ -1302,7 +1302,7 @@ uuid = "295af30f-e4ad-537b-8983-00126c2a3abe"
version = "3.5.18"
[[deps.Ribasim]]
-deps = ["Accessors", "Arrow", "BasicModelInterface", "CodecZstd", "ComponentArrays", "Configurations", "DBInterface", "DataInterpolations", "DataStructures", "Dates", "DiffEqCallbacks", "EnumX", "FiniteDiff", "ForwardDiff", "Graphs", "HiGHS", "IterTools", "JuMP", "Legolas", "LinearSolve", "Logging", "LoggingExtras", "MetaGraphsNext", "OrdinaryDiffEq", "PreallocationTools", "SQLite", "SciMLBase", "SparseArrays", "SparseConnectivityTracer", "StructArrays", "Tables", "TerminalLoggers", "TranscodingStreams"]
+deps = ["Accessors", "Arrow", "BasicModelInterface", "CodecZstd", "ComponentArrays", "Configurations", "DBInterface", "DataInterpolations", "DataStructures", "Dates", "DiffEqCallbacks", "EnumX", "FiniteDiff", "ForwardDiff", "Graphs", "HiGHS", "IterTools", "JuMP", "Legolas", "LineSearches", "LinearSolve", "Logging", "LoggingExtras", "MetaGraphsNext", "OrdinaryDiffEq", "PreallocationTools", "SQLite", "SciMLBase", "SparseArrays", "SparseConnectivityTracer", "StructArrays", "Tables", "TerminalLoggers", "TranscodingStreams"]
path = "core"
uuid = "aac5e3d9-0b8f-4d4f-8241-b1a7a9632635"
version = "2024.10.0"
@@ -1669,12 +1669,6 @@ weakdeps = ["RecipesBase"]
[deps.TimeZones.extensions]
TimeZonesRecipesBaseExt = "RecipesBase"
-[[deps.TimerOutputs]]
-deps = ["ExprTools", "Printf"]
-git-tree-sha1 = "5a13ae8a41237cff5ecf34f73eb1b8f42fff6531"
-uuid = "a759f4b9-e2f1-59dc-863e-4aeb61b1ea8f"
-version = "0.5.24"
-
[[deps.TranscodingStreams]]
git-tree-sha1 = "d73336d81cafdc277ff45558bb7eaa2b04a8e472"
uuid = "3bb67fe8-82b1-5028-8e26-92a6c54297fa"
diff --git a/core/Project.toml b/core/Project.toml
index ce3e4882c..cf25cc035 100644
--- a/core/Project.toml
+++ b/core/Project.toml
@@ -24,6 +24,7 @@ HiGHS = "87dc4568-4c63-4d18-b0c0-bb2238e4078b"
IterTools = "c8e1da08-722c-5040-9ed9-7db0dc04731e"
JuMP = "4076af6c-e467-56ae-b986-b466b2749572"
Legolas = "741b9549-f6ed-4911-9fbf-4a1c0c97f0cd"
+LineSearches = "d3d80556-e9d4-5f37-9878-2ab0fcc64255"
LinearSolve = "7ed4a6bd-45f5-4d41-b270-4a48e9bafcae"
Logging = "56ddb016-857b-54e1-b83d-db4d58db5568"
LoggingExtras = "e6f89c97-d47a-5376-807f-9c37f3926c36"
@@ -70,6 +71,7 @@ IOCapture = "0.2"
IterTools = "1.4"
JuMP = "1.15"
Legolas = "0.5"
+LineSearches = "7"
LinearSolve = "2.24"
Logging = "<0.0.1, 1"
LoggingExtras = "1"
diff --git a/core/ext/RibasimMakieExt.jl b/core/ext/RibasimMakieExt.jl
index 97077bab8..7c21330ae 100644
--- a/core/ext/RibasimMakieExt.jl
+++ b/core/ext/RibasimMakieExt.jl
@@ -1,13 +1,13 @@
module RibasimMakieExt
using DataFrames: DataFrame
-using Makie: Figure, Axis, lines!, axislegend
+using Makie: Figure, Axis, scatterlines!, axislegend
using Ribasim: Ribasim, Model
function Ribasim.plot_basin_data!(model::Model, ax::Axis, column::Symbol)
basin_data = DataFrame(Ribasim.basin_table(model))
for node_id in unique(basin_data.node_id)
group = filter(:node_id => ==(node_id), basin_data)
- lines!(ax, group.time, getproperty(group, column); label = "Basin #$node_id")
+ scatterlines!(ax, group.time, getproperty(group, column); label = "Basin #$node_id")
end
axislegend(ax)
@@ -23,31 +23,23 @@ function Ribasim.plot_basin_data(model::Model)
f
end
-function Ribasim.plot_flow!(
- model::Model,
- ax::Axis,
- edge_id::Int32;
- skip_conservative_out = false,
-)
+function Ribasim.plot_flow!(model::Model, ax::Axis, edge_metadata::Ribasim.EdgeMetadata)
flow_data = DataFrame(Ribasim.flow_table(model))
- flow_data = filter(:edge_id => ==(edge_id), flow_data)
- first_row = first(flow_data)
- # Skip outflows of conservative nodes because these are the same as the inflows
- if skip_conservative_out &&
- Ribasim.NodeType.T(first_row.from_node_type) in Ribasim.conservative_nodetypes
- return nothing
- end
- label = "$(first_row.from_node_type) #$(first_row.from_node_id) → $(first_row.to_node_type) #$(first_row.to_node_id)"
- lines!(ax, flow_data.time, flow_data.flow_rate; label)
+ flow_data = filter(:edge_id => ==(edge_metadata.id), flow_data)
+ label = "$(edge_metadata.edge[1]) → $(edge_metadata.edge[2])"
+ scatterlines!(ax, flow_data.time, flow_data.flow_rate; label)
return nothing
end
-function Ribasim.plot_flow(model::Model)
+function Ribasim.plot_flow(model::Model; skip_conservative_out = true)
f = Figure()
ax = Axis(f[1, 1]; xlabel = "time", ylabel = "flow rate [m³s⁻¹]")
- edge_ids = unique(Ribasim.flow_table(model).edge_id)
- for edge_id in edge_ids
- Ribasim.plot_flow!(model, ax, edge_id; skip_conservative_out = true)
+ for edge_metadata in values(model.integrator.p.graph.edge_data)
+ if skip_conservative_out &&
+ edge_metadata.edge[1].type in Ribasim.conservative_nodetypes
+ continue
+ end
+ Ribasim.plot_flow!(model, ax, edge_metadata)
end
axislegend(ax)
f
diff --git a/core/src/Ribasim.jl b/core/src/Ribasim.jl
index 059a62d4e..01d735dd2 100644
--- a/core/src/Ribasim.jl
+++ b/core/src/Ribasim.jl
@@ -15,7 +15,15 @@ For more granular access, see:
module Ribasim
# Algorithms for solving ODEs.
-using OrdinaryDiffEq: OrdinaryDiffEq, OrdinaryDiffEqRosenbrockAdaptiveAlgorithm, get_du
+using OrdinaryDiffEq:
+ OrdinaryDiffEq,
+ OrdinaryDiffEqRosenbrockAdaptiveAlgorithm,
+ get_du,
+ AbstractNLSolver,
+ relax!,
+ _compute_rhs!,
+ calculate_residuals!
+using LineSearches: BackTracking
# Interface for defining and solving the ODE problem of the physical layer.
using SciMLBase:
@@ -31,7 +39,8 @@ using SciMLBase:
ODEProblem,
ODESolution,
VectorContinuousCallback,
- get_proposed_dt
+ get_proposed_dt,
+ DEIntegrator
# Automatically detecting the sparsity pattern of the Jacobian of water_balance!
# through operator overloading
diff --git a/core/src/config.jl b/core/src/config.jl
index 794a17477..bb024cd0f 100644
--- a/core/src/config.jl
+++ b/core/src/config.jl
@@ -230,7 +230,7 @@ const algorithms = Dict{String, Type}(
)
"Create an OrdinaryDiffEqAlgorithm from solver config"
-function algorithm(solver::Solver)::OrdinaryDiffEqAlgorithm
+function algorithm(solver::Solver; u0 = [])::OrdinaryDiffEqAlgorithm
algotype = get(algorithms, solver.algorithm, nothing)
if algotype === nothing
options = join(keys(algorithms), ", ")
@@ -239,7 +239,9 @@ function algorithm(solver::Solver)::OrdinaryDiffEqAlgorithm
end
kwargs = Dict{Symbol, Any}()
if algotype <: OrdinaryDiffEqNewtonAdaptiveAlgorithm
- kwargs[:nlsolve] = NLNewton(; relax = 0.1)
+ kwargs[:nlsolve] = NLNewton(;
+ relax = Ribasim.MonitoredBackTracking(; z_tmp = copy(u0), dz_tmp = copy(u0)),
+ )
end
# not all algorithms support this keyword
kwargs[:autodiff] = solver.autodiff
diff --git a/core/src/model.jl b/core/src/model.jl
index 13d898336..9c6a447be 100644
--- a/core/src/model.jl
+++ b/core/src/model.jl
@@ -37,7 +37,6 @@ function Model(config_path::AbstractString)::Model
end
function Model(config::Config)::Model
- alg = algorithm(config.solver)
db_path = input_path(config, config.database)
if !isfile(db_path)
@error "Database file not found" db_path
@@ -109,6 +108,9 @@ function Model(config::Config)::Model
u0 = ComponentVector{Float64}(; storage, integral)
du0 = zero(u0)
+ # The Solver algorithm
+ alg = algorithm(config.solver; u0)
+
# Synchronize level with storage
set_current_basin_properties!(parameters.basin, u0, du0)
diff --git a/core/src/read.jl b/core/src/read.jl
index e76793e28..05102d64e 100644
--- a/core/src/read.jl
+++ b/core/src/read.jl
@@ -572,7 +572,8 @@ function Basin(db::DB, config::Config, graph::MetaGraph)::Basin
error("Invalid Basin / profile table.")
end
- level_to_area = LinearInterpolation.(area, level; extrapolate = true)
+ level_to_area =
+ LinearInterpolation.(area, level; extrapolate = true, cache_parameters = true)
storage_to_level = invert_integral.(level_to_area)
t_end = seconds_since(config.endtime, config.starttime)
@@ -921,6 +922,7 @@ function user_demand_static!(
fill(first_row.return_factor, 2),
return_factor_old.t;
extrapolate = true,
+ cache_parameters = true,
)
min_level[user_demand_idx] = first_row.min_level
@@ -1026,8 +1028,10 @@ function UserDemand(db::DB, config::Config, graph::MetaGraph)::UserDemand
]
demand_from_timeseries = fill(false, n_user)
allocated = fill(Inf, n_user, n_priority)
- return_factor =
- [LinearInterpolation(zeros(2), trivial_timespan) for i in eachindex(node_ids)]
+ return_factor = [
+ LinearInterpolation(zeros(2), trivial_timespan; cache_parameters = true) for
+ i in eachindex(node_ids)
+ ]
min_level = zeros(n_user)
# Process static table
diff --git a/core/src/solve.jl b/core/src/solve.jl
index cabfcd6e0..04aea58a7 100644
--- a/core/src/solve.jl
+++ b/core/src/solve.jl
@@ -51,9 +51,20 @@ function water_balance!(
# Formulate du (controlled by PidControl)
formulate_du_pid_controlled!(du, graph, pid_control)
+ # https://github.com/Deltares/Ribasim/issues/1705#issuecomment-2283293974
+ stop_declining_negative_storage!(du, u)
+
return nothing
end
+function stop_declining_negative_storage!(du, u)
+ for (i, s) in enumerate(u.storage)
+ if s < 0
+ du.storage[i] = max(du.storage[i], 0.0)
+ end
+ end
+end
+
function formulate_continuous_control!(du, p, t)::Nothing
(; compound_variable, target_ref, func) = p.continuous_control
diff --git a/core/src/util.jl b/core/src/util.jl
index 695f62435..90d197cf3 100644
--- a/core/src/util.jl
+++ b/core/src/util.jl
@@ -53,9 +53,18 @@ end
Compute the area and level of a basin given its storage.
"""
function get_area_and_level(basin::Basin, state_idx::Int, storage::T)::Tuple{T, T} where {T}
- level = basin.storage_to_level[state_idx](max(storage, 0.0))
- area = basin.level_to_area[state_idx](level)
-
+ storage_to_level = basin.storage_to_level[state_idx]
+ level_to_area = basin.level_to_area[state_idx]
+ if storage >= 0
+ level = storage_to_level(storage)
+ else
+ # Negative storage is not feasible and this yields a level
+ # below the basin bottom, but this does yield usable gradients
+ # for the non-linear solver
+ bottom = first(level_to_area.t)
+ level = bottom + derivative(storage_to_level, 0.0) * storage
+ end
+ area = level_to_area(level)
return area, level
end
@@ -887,3 +896,70 @@ end
(A::AbstractInterpolation)(t::GradientTracer) = t
reduction_factor(x::GradientTracer, threshold::Real) = x
relaxed_root(x::GradientTracer, threshold::Real) = x
+get_area_and_level(basin::Basin, state_idx::Int, storage::GradientTracer) = storage, storage
+stop_declining_negative_storage!(du, u::ComponentVector{<:GradientTracer}) = nothing
+
+@kwdef struct MonitoredBackTracking{B, V}
+ linesearch::B = BackTracking()
+ dz_tmp::V = []
+ z_tmp::V = []
+end
+
+"""
+Compute the residual of the non-linear solver, i.e. a measure of the
+error in the solution to the implicit equation defined by the solver algorithm
+"""
+function residual(z, integrator, nlsolver, f)
+ (; uprev, t, p, dt, opts, isdae) = integrator
+ (; tmp, ztmp, γ, α, cache, method) = nlsolver
+ (; ustep, atmp, tstep, k, invγdt, tstep, k, invγdt) = cache
+ if isdae
+ _uprev = get_dae_uprev(integrator, uprev)
+ b, ustep2 =
+ _compute_rhs!(tmp, ztmp, ustep, α, tstep, k, invγdt, p, _uprev, f::TF, z)
+ else
+ b, ustep2 =
+ _compute_rhs!(tmp, ztmp, ustep, γ, α, tstep, k, invγdt, method, p, dt, f, z)
+ end
+ calculate_residuals!(
+ atmp,
+ b,
+ uprev,
+ ustep2,
+ opts.abstol,
+ opts.reltol,
+ opts.internalnorm,
+ t,
+ )
+ ndz = opts.internalnorm(atmp, t)
+ return ndz
+end
+
+"""
+MonitoredBackTracing is a thin wrapper of BackTracking, making sure that
+the BackTracking relaxation is rejected if it results in a residual increase
+"""
+function OrdinaryDiffEq.relax!(
+ dz,
+ nlsolver::AbstractNLSolver,
+ integrator::DEIntegrator,
+ f,
+ linesearch::MonitoredBackTracking,
+)
+ (; linesearch, dz_tmp, z_tmp) = linesearch
+
+ # Store step before relaxation
+ @. dz_tmp = dz
+
+ # Apply relaxation and measure the residual change
+ @. z_tmp = nlsolver.z + dz
+ resid_before = residual(z_tmp, integrator, nlsolver, f)
+ relax!(dz, nlsolver, integrator, f, linesearch)
+ @. z_tmp = nlsolver.z + dz
+ resid_after = residual(z_tmp, integrator, nlsolver, f)
+
+ # If the residual increased due to the relaxation, reject it
+ if resid_after > resid_before
+ @. dz = dz_tmp
+ end
+end
diff --git a/core/test/main_test.jl b/core/test/main_test.jl
index 788e9c7c3..95dfac40e 100644
--- a/core/test/main_test.jl
+++ b/core/test/main_test.jl
@@ -24,7 +24,6 @@
@show backtrace
end
@test occursin("version in the TOML config file does not match", output)
- @test occursin("Info: Convergence bottlenecks in descending order of severity:", output)
end
@testitem "main error logging" begin