diff --git a/.github/workflows/doc_deploy.yml b/.github/workflows/doc_deploy.yml
new file mode 100644
index 00000000..b9c4034c
--- /dev/null
+++ b/.github/workflows/doc_deploy.yml
@@ -0,0 +1,27 @@
+# Deploy documentation when push to main
+
+name: Deploy docs
+
+on:
+ push:
+ branches: [ "main" ]
+
+jobs:
+ build:
+ name: Deploy docs
+ runs-on: ubuntu-latest
+ steps:
+ - name: Checkout main
+ uses: actions/checkout@v3
+ with:
+ fetch-depth: 0
+ - name: Set up Python 3.11
+ uses: actions/setup-python@v2
+ with:
+ python-version: "3.11"
+ - name: Install dependencies
+ run: |
+ python -m pip install --upgrade pip
+ python -m pip install -r docs/requirements.txt
+ - name: Deploy docs
+ run: mkdocs gh-deploy --force
diff --git a/CHANGELOG.md b/CHANGELOG.md
index e4b2edb2..20606166 100644
--- a/CHANGELOG.md
+++ b/CHANGELOG.md
@@ -1,5 +1,10 @@
# Unreleased
+**Added:**
+
+- Documentation using mkdocs and a github action workflow to publish the
+ documentation in [#264](https://github.com/EcoExtreML/STEMMUS_SCOPE/pull/264)
+
**Changed:**
@@ -50,7 +55,7 @@ as well as various bugfixes.
- Defining the indices of the first four layers discussed in
[#237](https://github.com/EcoExtreML/STEMMUS_SCOPE/issues/237) and fixed in
[#238](https://github.com/EcoExtreML/STEMMUS_SCOPE/pull/238)
-
+
# [1.5.0](https://github.com/EcoExtreML/STEMMUS_SCOPE/releases/tag/1.5.0) - 3 Jan 2024
diff --git a/CONTRIBUTING.md b/CONTRIBUTING.md
deleted file mode 100644
index 0ca2d975..00000000
--- a/CONTRIBUTING.md
+++ /dev/null
@@ -1,208 +0,0 @@
-# Contributing guidelines
-
-This repository includes the MATLAB source code of the STEMMUS-SCOPE model. We welcome any
-kind of contributions to our software, from simple comments or questions to a full
-[pull request](https://help.github.com/articles/about-pull-requests/). Please
-read and follow our contributing guidelines.
-
-## Contributing via GitHub
-
-If you want to work with the `STEMMUS_SCOPE` repository for the first time, or on a new computer,
-you need to configure a few things following steps 1 through 5 below.
-
-
- Steps 1 to 5
-
-### 1. Enable two-factor authentication
-
-It is strongly recommended using two-factor authentication. Here is the link of
-[Configuring two-factor
-authentication](https://docs.github.com/en/authentication/securing-your-account-with-two-factor-authentication-2fa/configuring-two-factor-authentication).
-
-### 2. Set ssh connection
-
-With SSH keys, you can connect to GitHub without supplying your username and
-personal access token at each visit. Please follow the instructions below. If
-you like to know more, see [Connecting to GitHub with
-SSH](https://docs.github.com/en/authentication/connecting-to-github-with-ssh)
-
-#### 2.1. Checking for existing SSH keys
-
-Open a terminal and run the command below:
-
-```bash
-ls -la ~/.ssh
-```
-
-This command lists the files with extension `.pub` like `id_rsa.pub` in the
-`.ssh` directory, if they exist. If you receive an error that `~/.ssh` doesn't
-exist, or you don't see any files with extension `.pub`, you do not have an
-existing SSH key pair. So, continue with step **2.2**. Otherwise, skip step 2.2 and
-continue with step **2.3**.
-
-#### 2.2. Generating a new SSH key
-
-Open a terminal and run the command below but replace `your_user_email` with
-your own GitHub email address:
-
-```ssh
-ssh-keygen -t ed25519 -C "your_user_email"
-```
-
-When you're prompted to "Enter a file in which to save the key," press `Enter`.
-This accepts the default file location.
-
-The next prompt asks "Enter passphrase (empty for no passphrase)", type a secure
-passphrase. For more information, see [Working with SSH key
-passphrases](https://docs.github.com/en/articles/working-with-ssh-key-passphrases).
-
-#### 2.3. Adding your SSH key to the ssh-agent
-
-Open a terminal and run the command below:
-
-```bash
-eval "$(ssh-agent -s)"
-```
-
-Then, run the command below:
-
-```bash
-ssh-add ~/.ssh/id_ed25519
-```
-
-This asks for your "passphrase" that was provided in the previous step.
-
-#### 2.4. Adding a new SSH key to your GitHub account
-
-Please follow steps 1 to 8 in this [GitHub
-instruction](https://docs.github.com/en/authentication/connecting-to-github-with-ssh/adding-a-new-ssh-key-to-your-github-account).
-
-### 3. Configure git
-
-#### 3.1. Set name and email
-
-Open a terminal, and run the commands below one by one but replace
-`your_user_name` and `your_user_email` with your own GitHub information:
-
-```bash
-git config --global user.name "your_user_name"
-git config --global user.email "your_user_email"
-```
-
-#### 3.2. Set line endings
-
-Change the way Git encodes line endings on Linux as:
-
-```bash
-git config --global core.autocrlf input
-```
-
-#### 3.3. Set text editor
-
-We can set `nano` as our favorite text editor, following:
-
-```bash
-git config --global core.editor "nano -w"
-```
-
-> We use `nano` here because it is one of the least complex text editors. Press
-> `ctrl + O` to save the file, and then `ctrl + X` to exit `nano`.
-
-#### 3.4. Check your settings
-
-You can check your settings at any time:
-
-```bash
-git config --list
-```
-
-For more information, see lesson [Setting Up
-Git](https://swcarpentry.github.io/git-novice/02-setup/index.html).
-
-### 4. Clone the repository
-
-Open a terminal and run the command below:
-
-```bash
-cd
-```
-Now you are in your `HOME` directory. Run the command below:
-
-```bash
-git clone git@github.com:EcoExtreML/STEMMUS_SCOPE.git
-```
-
-Now a new GitHub folder `STEMMUS_SCOPE` is created in your `HOME` directory.
-
-> In this command, we clone the repository using `ssh` option. As we set the ssh
-connection in [**Step 2**](#2-set-ssh-connection), this command here does not
-ask for our user name and password.
-
-### 5. Collaborate using GitHub
-
-To know about the most common Git commands, follow the guides
-[here](https://hackmd.io/B4v6KwsBRzG-akLDF8e5pg).
-
-
-## Development of the MATLAB source of STEMMUS_SCOPE model
-
-To contribute to the STEMMUS_SCOPE model, you need access to the model source code that is stored in the repository [STEMMUS_SCOPE](https://github.com/EcoExtreML/STEMMUS_SCOPE). You also need a MATLAB license. MATLAB `2021a` is installed on
-[Snellius]((https://servicedesk.surfsara.nl/wiki/display/WIKI/Snellius)) and [CRIB](https://crib.utwente.nl/), see [this instruction](https://pystemmusscope.readthedocs.io/en/latest/contributing_link.html#development-of-stemmus-scope-model).
-
-## Create an executable file of STEMMUS_SCOPE
-
-See the [exe readme](./exe/README.md).
-
-## Follow MATLAB style guidelines
-
-When you are introducing new changes to the codes, please follow the style introduced in [MATLAB Guidelines 2.0, Richard Johnson](http://cnl.sogang.ac.kr/cnlab/lectures/programming/matlab/Richard_Johnson-MatlabStyle2_book.pdf).
-
-When you submit a pull request, the code is also [checked](https://github.com/EcoExtreML/STEMMUS_SCOPE/actions/workflows/lint.yml) by the [MISS_HIT](misshit.org/) linter and style checker.
-The status of `MISS_HIT` checks is shown below the pull request. The checks should be successful (green) before merging the pull request.
-MISS_HIT is configured in [`miss_hit.cfg`](./miss_hit.cfg).
-
-### Installing MISS_HIT
-It is best practice to install packages in environments. See the dropdown menu for instructions.
-However, you can also continue below to install MISS_HIT without these steps.
-
-Python environment / conda instructions
-
-You need to have a valid python installation on your system.
-
-Create an enviroment with `venv` or conda:
-
-### **Venv**
-```bash
-python3 -m venv misshit # python on windows.
-```
-Activate this environment
-```bash
-source misshit/bin/activate
-```
-
-Or on Windows:
-```pwsh
-./misshit/Scripts/Activate.ps1
-```
-
-### **conda**
-```bash
-conda env create --name misshit
-conda activate misshit
-```
-
-
+ Steps 1 to 5
+
+### 1. Enable two-factor authentication
+
+It is strongly recommended using two-factor authentication. Here is the link of
+[Configuring two-factor
+authentication](https://docs.github.com/en/authentication/securing-your-account-with-two-factor-authentication-2fa/configuring-two-factor-authentication).
+
+### 2. Set ssh connection
+
+With SSH keys, you can connect to GitHub without supplying your username and
+personal access token at each visit. Please follow the instructions below. If
+you like to know more, see [Connecting to GitHub with
+SSH](https://docs.github.com/en/authentication/connecting-to-github-with-ssh)
+
+#### 2.1. Checking for existing SSH keys
+
+Open a terminal and run the command below:
+
+```bash
+ls -la ~/.ssh
+```
+
+This command lists the files with extension `.pub` like `id_rsa.pub` in the
+`.ssh` directory, if they exist. If you receive an error that `~/.ssh` doesn't
+exist, or you don't see any files with extension `.pub`, you do not have an
+existing SSH key pair. So, continue with step **2.2**. Otherwise, skip step 2.2 and
+continue with step **2.3**.
+
+#### 2.2. Generating a new SSH key
+
+Open a terminal and run the command below but replace `your_user_email` with
+your own GitHub email address:
+
+```ssh
+ssh-keygen -t ed25519 -C "your_user_email"
+```
+
+When you're prompted to "Enter a file in which to save the key," press `Enter`.
+This accepts the default file location.
+
+The next prompt asks "Enter passphrase (empty for no passphrase)", type a secure
+passphrase. For more information, see [Working with SSH key
+passphrases](https://docs.github.com/en/articles/working-with-ssh-key-passphrases).
+
+#### 2.3. Adding your SSH key to the ssh-agent
+
+Open a terminal and run the command below:
+
+```bash
+eval "$(ssh-agent -s)"
+```
+
+Then, run the command below:
+
+```bash
+ssh-add ~/.ssh/id_ed25519
+```
+
+This asks for your "passphrase" that was provided in the previous step.
+
+#### 2.4. Adding a new SSH key to your GitHub account
+
+Please follow steps 1 to 8 in this [GitHub
+instruction](https://docs.github.com/en/authentication/connecting-to-github-with-ssh/adding-a-new-ssh-key-to-your-github-account).
+
+### 3. Configure git
+
+#### 3.1. Set name and email
+
+Open a terminal, and run the commands below one by one but replace
+`your_user_name` and `your_user_email` with your own GitHub information:
+
+```bash
+git config --global user.name "your_user_name"
+git config --global user.email "your_user_email"
+```
+
+#### 3.2. Set line endings
+
+Change the way Git encodes line endings on Linux as:
+
+```bash
+git config --global core.autocrlf input
+```
+
+#### 3.3. Set text editor
+
+We can set `nano` as our favorite text editor, following:
+
+```bash
+git config --global core.editor "nano -w"
+```
+
+> We use `nano` here because it is one of the least complex text editors. Press
+> `ctrl + O` to save the file, and then `ctrl + X` to exit `nano`.
+
+#### 3.4. Check your settings
+
+You can check your settings at any time:
+
+```bash
+git config --list
+```
+
+For more information, see lesson [Setting Up
+Git](https://swcarpentry.github.io/git-novice/02-setup/index.html).
+
+### 4. Clone the repository
+
+Open a terminal and run the command below:
+
+```bash
+cd
+```
+Now you are in your `HOME` directory. Run the command below:
+
+```bash
+git clone git@github.com:EcoExtreML/STEMMUS_SCOPE.git
+```
+
+Now a new GitHub folder `STEMMUS_SCOPE` is created in your `HOME` directory.
+
+> In this command, we clone the repository using `ssh` option. As we set the ssh
+connection in [**Step 2**](#2-set-ssh-connection), this command here does not
+ask for our user name and password.
+
+### 5. Collaborate using GitHub
+
+To know about the most common Git commands, follow the guides
+[here](https://hackmd.io/B4v6KwsBRzG-akLDF8e5pg).
+
+
+## Adding changes to the model source code
+
+To setup the required software and configurations, see the documentation on
+["**Getting started**"](./getting_started.md).
+
+It would be ideal to introduce changes incrementally over time. This way, you
+can track the changes and understand the impact of each change. Here are the
+steps to follow:
+
+- Submit [an issue](https://github.com/EcoExtreML/STEMMUS_SCOPE/issues) to the
+ repository. This issue should describe the problem or the feature you want to
+ introduce.
+- Create a new branch from the `main` branch. This branch should have a
+ descriptive name that relates to the issue you are working on.
+- Make the changes in the new branch, for example, you can add new features, fix
+ bugs, or improve the documentation. Follow [MATLAB Guidelines 2.0, Richard
+ Johnson](http://cnl.sogang.ac.kr/cnlab/lectures/programming/matlab/Richard_Johnson-MatlabStyle2_book.pdf)
+ when introducing new changes to the codes.
+- Commit the changes to the new branch. Write a descriptive commit message that
+ explains the changes you are introducing.
+- Push the changes to the repository.
+- Create a **draft** [pull
+ request](https://docs.github.com/en/pull-requests/collaborating-with-pull-requests/proposing-changes-to-your-work-with-pull-requests/about-pull-requests)
+ to the `main` branch. This pull request should reference the issue you are
+ working on.
+- When you are done with the changes, run [the tests](#testing-new-changes) and
+ [MISS_HIT commands](#miss_hit-checks), make sure they pass.
+- Make the pull request ready for review. Ask for a review from the repository
+ maintainers.
+- The maintainers will review the changes and provide feedback. You may need to
+ make additional changes based on the feedback.
+- When the changes are approved, the maintainers will merge the pull request.
+
+## Reviewing a pull request
+
+When you are reviewing a pull request, you should follow the steps below:
+
+- Check the changes introduced in the pull request. Make sure the changes are
+ consistent with the issue description. Also, check for example variable names,
+ function names, documentation, global variables, ..., to see if the changes
+ are consistent with the [MATLAB Style Guidelines
+ 2.0](http://cnl.sogang.ac.kr/cnlab/lectures/programming/matlab/Richard_Johnson-MatlabStyle2_book.pdf).
+- Run [the tests](#testing-new-changes) and [MISS_HIT commands](#miss_hit-checks) to make sure they pass.
+- Provide feedback on the changes. You can ask questions, suggest improvements,
+ or point out issues.
+- When you are satisfied with the changes, approve the pull request. Ask a
+ maintainer to [re-generate the executable
+ file](#creating-an-executable-file-of-stemmus_scope) and merge the pull
+ request.
+- If the changes are related to BMI, you have to ask the maintainer to make a
+ new release of the model. This way a new docker image will be created and the
+ model will be available for the users.
+- If the changes are related to the documentation, make sure that documentation
+ page can be built successfully, see [Building the documentation
+ locally](#building-the-documentation-locally).
+
+## Merging a pull request
+
+When you are merging a pull request, you should follow the steps below:
+
+- Make sure the pull request is approved by at least one reviewer.
+- Make sure all the items in the pull request's list are checked.
+
+## Creating an executable file of STEMMUS_SCOPE
+
+See the disumentation on [executable
+file](https://github.com/EcoExtreML/STEMMUS_SCOPE/tree/main/run_model_on_snellius/exe).
+
+## MISS_HIT Checks
+
+When you submit a pull request, the code is also
+[checked](https://github.com/EcoExtreML/STEMMUS_SCOPE/actions/workflows/lint.yml)
+by the [MISS_HIT](https://misshit.org/) linter and style checker. The status of
+`MISS_HIT` checks is shown below the pull request. The checks should be
+successful (green) before merging the pull request. To work with `MISS_HIT`,
+follow the instructions below:
+
+- Installing MISS_HIT: To install MISS_HIT, follow their [installation
+ instructions](https://github.com/florianschanda/miss_hit#installation-via-pip).
+
+- Running MISS_HIT: To run the style checker or linter, navigate to the `src/`
+ folder in STEMMUS_SCOPE, and run the following commands:
+
+ ```bash
+ mh_style
+ mh_lint
+ mh_metric
+ ```
+
+ Follow the errors and information in the output to fix any issues.
+
+- Configuring MISS_HIT: `MISS_HIT` is configured in
+[`miss_hit.cfg`](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/miss_hit.cfg).
+This file contains the configuration for the linter and style checker. Please do
+not change this file unless you are familiar with the configuration options.
+
+## Testing new changes
+
+To test the new changes, we can only check if the new changes do not break the
+model. To do this, follow the steps below:
+
+- Run the notebook `compare_git_branch_results.ipynb` in the `test` folder. This
+ notebook will compare the results of the model between the `main` branch and
+ the new branch with the changes. If the results are similar, the changes do
+ not break the model.
+- [Create the executable file](#creating-an-executable-file-of-stemmus_scope) of
+ the model and run the model with the new changes. Check if the model runs
+ without any errors.
+- [Run the model with Octave](#octave-compatibility) to check if the new changes
+ are compatible with Octave.
+
+## Building the documentation locally
+
+Documentation is created using several markdown files and the [`mkdocs`
+tool](https://www.mkdocs.org/). The markdown files are located in the `docs`
+folder. The configuration file for `mkdocs` is `mkdocs.yml` located in the root
+directory. To build the documentation locally, follow the steps below:
+
+- Install the required dependencies as:
+
+ ```bash
+ cd STEMMUS_SCOPE
+ pip install -r docs/requirements.txt
+ ```
+
+- Build the documentation as:
+
+ ```bash
+ mkdocs build
+ ```
+
+- Preview the documentation as:
+
+ ```bash
+ mkdocs serve
+ ```
+
+Click on the link provided in the terminal to view the documentation in your
+browser.
+
+## Making a release
+
+When you are ready to make a release of the model, follow the steps below:
+
+- Make sure all new changes are added to changes log in the file
+ ["CHANGELOG.md"](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/CHANGELOG.md).
+- Create a new release in the repository. The release should have a version number
+ following the [semantic versioning](https://semver.org/) guidelines.
+- Add a description of the changes in the release notes.
+
+## GitHub actions workflow
+
+[GitHub
+actions](https://docs.github.com/en/actions/about-github-actions/understanding-github-actions)
+workflows are located in the folder `.github/workflows`:
+
+- `lint.yml`: This workflow checks the code style and lints the code using
+ `MISS_HIT`.
+- `doc_deploy.yml`: This workflow builds the documentation and deploys it to
+ GitHub pages. It will create a gh-pages branch in the repository.
+- `publish-container`: This workflow builds the docker image and publishes it to
+ the repository once a new release is made. The docker image will avialable in
+ the
+ [packages](https://github.com/EcoExtreML/STEMMUS_SCOPE/pkgs/container/stemmus_scope)
+ in the repository.
+
+## Docker image
+
+The
+[Dockerfile](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/Dockerfile)
+is located in the root directory. This file is used to build the docker image of
+the model by Github action `publish-container`, see [GitHub actions
+workflow](#github-actions-workflow). To build and run the docker image locally,
+follow the steps below:
+
+- Install [Docker](https://www.docker.com/).
+- Build the docker image **locally** as:
+
+ ```bash
+ cd STEMMUS_SCOPE
+ docker build -t stemmus_scope .
+ ```
+
+- Run the docker image with the BMI interface as:
+
+ ```bash
+ docker run -u $(id -u):$(id -g) -v /path_to_input_folder:/path_to_input_folder -v /path_to_output_folder:/path_to_output_folder -it stemmus_scope
+ ```
+
+ With `-it` option, you can enter the docker interactive mode.
+ Now, BMI asks you to select one of the steps "initialize", "update" or "finalize".
+ To initialize the model using a config file, type:
+
+ ```bash
+ initialize "path/to/config_file_template.txt"
+ ```
+
+## Octave compatibility
+
+See the documentation on [Octave](./Octave_instructions.md).
diff --git a/Octave_instructions.md b/docs/Octave_instructions.md
similarity index 61%
rename from Octave_instructions.md
rename to docs/Octave_instructions.md
index a951db9a..27d3fc0c 100644
--- a/Octave_instructions.md
+++ b/docs/Octave_instructions.md
@@ -1,26 +1,56 @@
-# Using STEMMUS-SCOPE with GNU Octave
+# Running STEMMUS-SCOPE in Octave
-- [Using STEMMUS-SCOPE with GNU Octave](#using-stemmus-scope-with-gnu-octave)
- - [VS Code setup](#vs-code-setup)
- - [Running STEMMUS-SCOPE in Octave](#running-stemmus-scope-in-octave)
- - [Developing STEMMUS-SCOPE in Octave](#developing-stemmus-scope-in-octave)
- - [Octave GUI](#octave-gui)
- - [VS Code](#vs-code)
- - [VS Code + Dev container](#vs-code--dev-container)
- - [Mount extra directory](#mount-extra-directory)
- - [Linux from source](#linux-from-source)
+If you want to run the model for a small time period or tests purpose, you
+can use [Octave](https://octave.org/). Allmost all funcationalities of
+STEMMUS_SCOPE are compatible with Octave, but the execution time is longer
+than MATLAB. After Octave installation, launch octave and install the following Octave packages:
-The downloads can be found here
-https://octave.org/download
+```bash
+pkg install "https://downloads.sourceforge.net/project/octave/Octave%20Forge%20Packages/Individual%20Package%20Releases/io-2.6.4.tar.gz"
+pkg install "https://downloads.sourceforge.net/project/octave/Octave%20Forge%20Packages/Individual%20Package%20Releases/statistics-1.4.3.tar.gz"
+```
+
+Then, pass config file path to the variable `CFG` in the main script
+`STEMMUS_SCOPE.m`. Run the model in a terminal:
+
+```bash
+cd src/
+octave.bat --no-gui --interactive --silent --eval "STEMMUS_SCOPE"
+```
+
+On a Unix system, use `octave` instead of `octave.bat`.
+
+
+Octave from source
+
+Note that Octave on many Linux distributions might be too old so we need to compile it ourselves.
+See [https://wiki.octave.org/Building](https://wiki.octave.org/Building). Here are build instructions for Ubuntu 22.04:
+
+```shell
+sudo apt update
+# install minimal deps, see https://wiki.octave.org/Octave_for_Debian_systems#The_right_way for all dependencies
+sudo apt install -yq wget build-essential gfortran liblapack-dev libblas-dev libpcre3-dev libreadline-dev libnetcdf-dev
+wget https://mirror.serverion.com/gnu/octave/octave-7.2.0.tar.gz # or download from local mirror at https://ftpmirror.gnu.org/octave
+tar -zxf octave-7.2.0.tar.gz
+cd octave-7.2.0
+./configure --prefix=/opt/octave
+make -j 6
+sudo make install
+```
-TODO: installation of octave on linux
-After installation, launch octave and install the following Octave package:
-`pkg install -forge statistics`
+Add `/opt/octave/bin` to PATH environment variable.
-For off-line installation, first, download the package [io](https://octave.sourceforge.io/io/index.html) and [statistics](https://octave.sourceforge.io/statistics/index.html). Then, launch octave and run:
+```shell
+export PATH=$PATH:/opt/octave/bin
+```
-`pkg install io-2.6.4.tar.gz`
-`pkg install statistics-1.4.3.tar.gz`
+Launch Octave and install Octave dependencies with:
+
+```bash
+pkg install "https://downloads.sourceforge.net/project/octave/Octave%20Forge%20Packages/Individual%20Package%20Releases/io-2.6.4.tar.gz"
+pkg install "https://downloads.sourceforge.net/project/octave/Octave%20Forge%20Packages/Individual%20Package%20Releases/statistics-1.4.3.tar.gz"
+```
+
## VS Code setup
Add Octave to path, e.g. for (64-bit) Windows add the following folders:
@@ -35,23 +65,18 @@ The debugger configurations are included in `/.vscode/launch.json`
`Octave Hacking` by Andrew Janke https://marketplace.visualstudio.com/items?itemName=apjanke.octave-hacking
This adds syntax highlighting and formatting.
-## Running STEMMUS-SCOPE in Octave
-It is possible to run STEMMUS-SCOPE from the command line with the following setup:
-`octave.bat --no-gui --interactive --silent --eval "STEMMUS_SCOPE_exe('path_to_config_file')"`
-
-On a Unix system, use `octave` instead of `octave.bat`.
## Developing STEMMUS-SCOPE in Octave
-Open the `run_Octave.m` file, either in VS Code or the Octave GUI.
+Open the `debug_Octave.m` file, either in VS Code or the Octave GUI.
### Octave GUI
-Set the workspace to the `STEMMUS_SCOPE/src` folder, and open the `run_Octave.m` file.
+Set the workspace to the `STEMMUS_SCOPE/src` folder, and open the `debug_Octave.m` file.
Here you can set the config file that should be used, and then run the file.
### VS Code
While having the `STEMMUS_SCOPE` folder as the workspace, open the debugger and select `Octave: Debug STEMMUS-SCOPE`.
Start the debugger to run (and debug) the model.
-In the `run_Octave.m` file you can set the config file that should be used.
+In the `debug_Octave.m` file you can set the config file that should be used.
## VS Code + Dev container
@@ -65,7 +90,10 @@ It will then start a Docker container with your code, Octave and the VS Code Oct
By default a Dev container only has the current VS code folder mounted inside the container.
-To add additional directories like directory with model input files to the container you will need to edit the [.devcontainer/devcontainer.json](.devcontainer/devcontainer.json) and add
+To add additional directories like directory with model input files to the
+container you will need to edit the
+[.devcontainer/devcontainer.json](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/.devcontainer/devcontainer.json)
+and add
```json
"mounts": [
@@ -78,36 +106,3 @@ After editing file you can restart the editor to get the extra directory inside
See [dev container docs](https://code.visualstudio.com/remote/advancedcontainers/add-local-file-mount) for more info.
To mount Windows directory inside the dev container you have to start the container in WSL2 (aka run Docker service inside WSL2) and use unix paths like `/mnt/c/...`.
-
-## Linux from source
-
-Octave on many Linux distributions is too old so we need to compile it ourselves.
-See [https://wiki.octave.org/Building](https://wiki.octave.org/Building).
-
-
-Here are build instructions for Ubuntu 22.04
-
-```shell
-sudo apt update
-# install minimal deps, see https://wiki.octave.org/Octave_for_Debian_systems#The_right_way for all dependencies
-sudo apt install -yq wget build-essential gfortran liblapack-dev libblas-dev libpcre3-dev libreadline-dev libnetcdf-dev
-wget https://mirror.serverion.com/gnu/octave/octave-7.2.0.tar.gz # or download from local mirror at https://ftpmirror.gnu.org/octave
-tar -zxf octave-7.2.0.tar.gz
-cd octave-7.2.0
-./configure --prefix=/opt/octave
-make -j 6
-sudo make install
-```
-
-Add `/opt/octave/bin` to PATH environment variable.
-
-```shell
-export PATH=$PATH:/opt/octave/bin
-```
-
-Install Octave dependencies with
-
-```shell
-octave --eval 'pkg install -forge statistics'
-```
-
\ No newline at end of file
diff --git a/docs/STEMMUS_SCOPE_BMI.md b/docs/STEMMUS_SCOPE_BMI.md
index 614e486b..362018be 100644
--- a/docs/STEMMUS_SCOPE_BMI.md
+++ b/docs/STEMMUS_SCOPE_BMI.md
@@ -13,7 +13,8 @@ When starting the executable, a run-mode can be specified. The following command
./STEMMUS_SCOPE "/home/path/to/config/file.txt" full
```
-(Where `./STEMMUS_SCOPE` is the path to the executable. For more info see [documentation](../run_model_on_snellius/exe/README.md))
+(Where `./STEMMUS_SCOPE` is the path to the executable. For more info see
+[documentation](https://github.com/EcoExtreML/STEMMUS_SCOPE/tree/main/run_model_on_snellius/exe))
To start BMI mode, pass anything (e.g. an empty string "") as config file, and use `bmi` to start the model in BMI-mode:
diff --git a/docs/STEMMUS_SCOPE_on_CRIB.md b/docs/STEMMUS_SCOPE_on_CRIB.md
index b35d0668..eb3d5c43 100644
--- a/docs/STEMMUS_SCOPE_on_CRIB.md
+++ b/docs/STEMMUS_SCOPE_on_CRIB.md
@@ -2,74 +2,85 @@
[CRIB](https://crib.utwente.nl/) is the ITC Geospatial Computing Platform.
-### Dataflow of STEMMUS_SCOPE on CRIB:
-
-1. Data required by the model are in a folder named "input" under project
- directory on CRIB. This folder includes:
-
- - Plumber2_data: the forcing/driving data provided by PLUMBER2.
- - SoilProperty: the soil texture data and soil hydraulic parameters.
-
- Below the directory explanations are from [SCOPE
- documentation](https://scope-model.readthedocs.io/en/latest/directories.html):
-
- - directional: the observer’s zenith and azimuth angles.(only used for
- multi-angle simulations (if the option ‘directional’ is switched on in
- parameters).
- - fluspect_parameters: absorption spectra of different leaf components are
- provided, according to PROSPECT 3.1, as well as Fluspect input: standard
- spectra for PSI and PSII.
- - leafangles: example leaf inclination distribution data are provided.
- - radiationdata: RTMo.m calculates spectra based on MODTRAN5 outputs (T-18
- system).Note that in the input data (files as well as the spreadsheet),
- the broadband input radiation may be provided. SCOPE linearly scales the
- input spectra of the optical and the thermal domain in such a way that
- the spectrally integrated input shortwave and long-wave radiation matches
- with the measured values.
- - soil_spectra: the soil spectrum is provided. Note that it is also possible
- to simulate a soil reflectance spectrum with the BSM model. In that case,
- the values for the BSM model parameters are taken from the input data, and
- the archived spectra in this folder are not used.
- - input_data.xlsx: the input to SCOPE model is provided. It
- provides parameter inputs for PROSPECT, leaf_biochemical, fluorescence,
- soil, canopy, aerodynamic, angles, photosynthetic temperature dependence
- functional parameters, etc.
- - input_soilLayThick.csv (optional): A file to change the discretization of
- the soil layers of the STEMMUS model. An example of this file is in
- [example_data folder](../example_data). This file (if needed) should be copied into the
- `InputPath` folder. If this file is used, it will override the default settings of
- the soil layers. The file has three columns: 1) layer number, 2) layer thickness,
- and 3) maximum root depth. The user is free to change the values of the three columns.
- Also, the number of rows determines the number of the soil layers and the total
- thickness of the soil column (sum of soil layer thickness).
-
-2. Config file: it is a text file that sets the paths **required** by the
- model. For example, see [config_file_crib.txt](../config_file_crib.txt) in this
- repository. This file includes:
-
- - SoilPropertyPath: a path to soil texture data and soil hydraulic
- parameters.
- - InputPath: this is the working/running directory of the model and should
- include the data of `directional`, `fluspect_parameters`, `leafangles`,
- `radiationdata`, `soil_spectra`, and `input_data.xlsx`.
- - OutputPath: this is the base path to store outputs of the model. When the
- model runs, it creates `sitename_timestamped` directories under this
- path.
- - ForcingPath: a path to the forcing/driving data. I.e. the Plumber2 dataset.
- - Location: Location where the model should be run. Currently,
- the model runs at the site scale. For example, if we put `FI-Hyy` here, the model
- runs at the `FI-Hyy` site.
- - StartTime: The start time of the model, in the ISO 8601 format. For example:
- `2001-01-01T00:00`. Note that the time can only be defined in half hour increments.
- If you want the start time to be the first available data point of the forcing data,
- you can set StartTime to `NA`.
- - EndTime: The end time of the model. Formatted the same way as the StartTime.
- For example: `2001-12-31T23:30`. If you want the end time to be the last available
- data point of the forcing data, you can set EndTime to `NA`.
-
- To edit the config file, open the file with a text editor and change the
- paths. The variable names e.g. `SoilPropertyPath` should not be changed.
- Also, note a `/` is required at the end of each line.
+### Dataflow of STEMMUS_SCOPE on CRIB
+
+To run the STEMMUS-SCOPE model, you need to have input data either from in-situ
+measurements or from remote sensing. Plumber2 site data are avialable under
+project directory on CRIB. This folder includes:
+
+- Plumber2_data: the forcing/driving data provided by PLUMBER2.
+- SoilProperty: the soil texture data and soil hydraulic parameters.
+
+In addition to site data, the remote sensing data are available on CRIB in
+`global_data` folder.
+
+Data required by the model are in a folder named "input". Below the directory
+explanations are from [SCOPE
+documentation](https://scope-model.readthedocs.io/en/latest/directories.html):
+
+- directional: the observer’s zenith and azimuth angles.(only used for
+ multi-angle simulations (if the option ‘directional’ is switched on in
+ parameters).
+- fluspect_parameters: absorption spectra of different leaf components are
+ provided, according to PROSPECT 3.1, as well as Fluspect input: standard
+ spectra for PSI and PSII.
+- leafangles: example leaf inclination distribution data are provided.
+- radiationdata: RTMo.m calculates spectra based on MODTRAN5 outputs (T-18
+ system).Note that in the input data (files as well as the spreadsheet),
+ the broadband input radiation may be provided. SCOPE linearly scales the
+ input spectra of the optical and the thermal domain in such a way that
+ the spectrally integrated input shortwave and long-wave radiation matches
+ with the measured values.
+- soil_spectra: the soil spectrum is provided. Note that it is also possible
+ to simulate a soil reflectance spectrum with the BSM model. In that case,
+ the values for the BSM model parameters are taken from the input data, and
+ the archived spectra in this folder are not used.
+- input_data.xlsx: the input to SCOPE model is provided. It
+ provides parameter inputs for PROSPECT, leaf_biochemical, fluorescence,
+ soil, canopy, aerodynamic, angles, photosynthetic temperature dependence
+ functional parameters, etc.
+- input_soilLayThick.csv (optional): A file to change the discretization of the
+ soil layers of the STEMMUS model. An example of this file is in [example_data
+ folder](https://github.com/EcoExtreML/STEMMUS_SCOPE/tree/main/example_data).
+ This file (if needed) should be copied into the
+ `InputPath` folder. If this file is used, it will override the default settings of
+ the soil layers. The file has three columns: 1) layer number, 2) layer thickness,
+ and 3) maximum root depth. The user is free to change the values of the three columns.
+ Also, the number of rows determines the number of the soil layers and the total
+ thickness of the soil column (sum of soil layer thickness).
+
+### Configuration file
+
+Config file: it is a text file that sets the paths **required** by the model.
+For example, see
+[config_file_crib.txt](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/config_file_crib.txt)
+or
+[config_file_crib_global.txt](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/config_file_crib_global.txt)
+in this repository. This file includes:
+
+- SoilPropertyPath: a path to soil texture data and soil hydraulic
+ parameters.
+- InputPath: this is the working/running directory of the model and should
+ include the data of `directional`, `fluspect_parameters`, `leafangles`,
+ `radiationdata`, `soil_spectra`, and `input_data.xlsx`.
+- OutputPath: this is the base path to store outputs of the model. When the
+model runs, it creates `sitename_timestamped` directories under this
+path.
+- ForcingPath: a path to the forcing/driving data. I.e. the Plumber2 dataset.
+- Location: Location where the model should be run. Currently,
+the model runs at the site scale. For example, if we put `FI-Hyy` here, the model
+runs at the `FI-Hyy` site.
+- StartTime: The start time of the model, in the ISO 8601 format. For example:
+`2001-01-01T00:00`. Note that the time can only be defined in half hour increments.
+If you want the start time to be the first available data point of the forcing data,
+you can set StartTime to `NA`.
+- EndTime: The end time of the model. Formatted the same way as the StartTime.
+For example: `2001-12-31T23:30`. If you want the end time to be the last available
+data point of the forcing data, you can set EndTime to `NA`.
+
+To edit the config file, open the file with a text editor and change the
+paths. The variable names e.g. `SoilPropertyPath` should not be changed.
+Also, note a `/` is required at the end of each line.
As explained above, the "InputPath" directory of the model is considered as
the working/running directory and should include some data required by the
@@ -80,56 +91,3 @@ different working/running directory and set the "InputPath" to it. Then,
you should copy the required data i.e. `directional`, `fluspect_parameters`,
`leafangles`, `radiationdata`, `soil_spectra`, and `input_data.xlsx` to the
working/running directory.
-
-### Workflow of STEMMUS_SCOPE on CRIB:
-
-1. The model reads the forcing file associated with the specified location,
- e.g., `FI-Hyy_1996-2014_FLUXNET2015_Met.nc` from "ForcingPath" and
- extracts forcing variables in `.dat` format using `filesread.m`.
- The `.dat` files are stored in the "InputPath" directory. In
- addition, the model reads the site information i.e. the location and
- vegetation parameters.
-1. The model reads the soil parameters from "SoilPropertyPath" using
- `soilpropertyread.m`.
-2. Some constants are loaded using `Constant.m`.
-3. The model runs step by step until the whole simulation period is completed
- i.e till the last time step of the forcing data.
-4. The results are saved as binary files temporarily. Then, the binary files are
- converted to `.csv` files and stored in a `sitename_timestamped` output
- directory under "OutputPath".
-
-### How to run STEMMUS_SCOPE on CRIB:
-
-1. Log in CRIB with your username and password and select a proper compute unit.
-2. Download the source code from this repository or get it using `git clone` in
- a terminal:
-
- ` git clone https://github.com/EcoExtreML/STEMMUS_SCOPE.git `
-
- All the codes can be found in the folder `src` whereas the executable file in
- the folder `exe`.
-
-3. Check `config_file_crib.txt` and change the paths if needed, specifically
- "InputPath" and "OutputPath".
-4. Follow the instructions below:
-
-#### Run using MATLAB that requires a license
-
-If you want to use MATLAB desktop,
-
-1. click on the `Remote Desktop` in the
-Launcher. Click on the `Applications`. You will find the 'MATLAB' software under
-the `Research`.
-2. After clicking on 'MATLAB', it asks for your account information that is
-connected to a MATLAB license.
-3. Open the file `filesread.m` and set the
-variable `CFG` to the path of the config file e.g. `CFG =
-'/data/shared/EcoExtreML/STEMMUS_SCOPEv1.0.0/STEMMUS_SCOPE/config_file_crib.txt';`.
-4. Then, run the main script `STEMMUS_SCOPE.m`.
-
-As an alternative, you can run the
-main script using MATLAB command line in a terminal:
-
-```bash
-matlab -nodisplay -nosplash -nodesktop -r "run('STEMMUS_SCOPE.m');exit;"
-```
diff --git a/docs/STEMMUS_SCOPE_on_Snellius.md b/docs/STEMMUS_SCOPE_on_Snellius.md
index 19525286..295e3074 100644
--- a/docs/STEMMUS_SCOPE_on_Snellius.md
+++ b/docs/STEMMUS_SCOPE_on_Snellius.md
@@ -3,54 +3,61 @@
[Snellius](https://servicedesk.surfsara.nl/wiki/display/WIKI/Snellius) is the
Dutch National supercomputer hosted at SURF.
-### Dataflow of STEMMUS_SCOPE on Snellius:
-
-1. Data required by the model are in a folder named "data" in the project
- directory `einf2480` on Snellius. This directory includes:
-
- - forcing/plumber2_data: the forcing/driving data provided by PLUMBER2.
- - model_parameters/soil_property: the soil texture data and soil hydraulic parameters.
- - model_parameters/vegetation_property:
- - directional
- - fluspect_parameters
- - leafangles
- - radiationdata
- - soil_spectra
- - input_data.xlsx
- - input_soilThick.csv (optional)
-
- For the explanation of the directories see
- [Dataflow of STEMMUS_SCOPE on CRIB](./STEMMUS_SCOPE_on_CRIB.md#dataflow-of-stemmus_scope-on-crib).
-
-2. Config file: it is a text file that sets the paths **required** by the model.
- For example, see [config_file_snellius.txt](../config_file_snellius.txt) in
- this repository. This file includes:
-
- - SoilPropertyPath: a path to soil texture data and soil hydraulic
- parameters.
- - InputPath: this is the working/running directory of the model and should
- include the data of `directional`, `fluspect_parameters`, `leafangles`,
- `radiationdata`, `soil_spectra`, and `input_data.xlsx`.
- - OutputPath: this is the base path to store outputs of the model. When the
- model runs, it creates `sitename_timestamped` directories under this
- path.
- - ForcingPath: a path to the forcing/driving data. I.e. the Plumber2 dataset.
- - Location: Location where the model should be run. Currently,
- the model runs at the site scale. For example, if we put `FI-Hyy` here, the model
- runs at the `FI-Hyy` site.
- - StartTime: The start time of the model, in the ISO 8601 format. For example:
- `2001-01-01T00:00`. Note that the time can only be defined in half hour increments.
- If you want the start time to be the first available data point of the forcing data,
- you can set StartTime to `NA`.
- - EndTime: The end time of the model. Formatted the same way as the StartTime.
- For example: `2001-12-31T23:30`. If you want the end time to be the last available
- data point of the forcing data, you can set EndTime to `NA`.
-
- To edit the config file, open the file with a text editor and change the
- paths. The `InputPath` and `OutputPath` are user-defined directories, make
- sure they exist and you have right permissions. The variable name e.g.
- `SoilPropertyPath` should not be changed. Also, note a `/` is required at
- the end of each line.
+### Dataflow of STEMMUS_SCOPE on Snellius
+
+To run the STEMMUS-SCOPE model, you need to have input data either from in-situ
+measurements or from remote sensing. Data required by the model are in a folder
+named "data" in the project directory `einf2480` on Snellius. This directory
+includes:
+
+- forcing/plumber2_data: the forcing/driving data provided by PLUMBER2.
+- model_parameters/soil_property: the soil texture data and soil hydraulic parameters.
+- model_parameters/vegetation_property:
+ - directional
+ - fluspect_parameters
+ - leafangles
+ - radiationdata
+ - soil_spectra
+ - input_data.xlsx
+ - input_soilThick.csv (optional)
+
+In addition to site data, the remote sensing data are available on Snellius in
+`global_data` folder.
+
+### Configuration file
+
+Config file: it is a text file that sets the paths **required** by the model.
+For example, see
+[config_file_snellius.txt](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/run_model_on_snellius/config_file_snellius.txt)
+or
+[config_file_snellius_global.txt](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/run_model_on_snellius/config_file_snellius_global.txt)
+in this repository. This file includes:
+
+- SoilPropertyPath: a path to soil texture data and soil hydraulic
+ parameters.
+- InputPath: this is the working/running directory of the model and should
+ include the data of `directional`, `fluspect_parameters`, `leafangles`,
+ `radiationdata`, `soil_spectra`, and `input_data.xlsx`.
+- OutputPath: this is the base path to store outputs of the model. When the
+model runs, it creates `sitename_timestamped` directories under this
+path.
+- ForcingPath: a path to the forcing/driving data. I.e. the Plumber2 dataset.
+- Location: Location where the model should be run. Currently,
+the model runs at the site scale. For example, if we put `FI-Hyy` here, the model
+runs at the `FI-Hyy` site.
+- StartTime: The start time of the model, in the ISO 8601 format. For example:
+`2001-01-01T00:00`. Note that the time can only be defined in half hour increments.
+If you want the start time to be the first available data point of the forcing data,
+you can set StartTime to `NA`.
+- EndTime: The end time of the model. Formatted the same way as the StartTime.
+For example: `2001-12-31T23:30`. If you want the end time to be the last available
+data point of the forcing data, you can set EndTime to `NA`.
+
+To edit the config file, open the file with a text editor and change the
+paths. The `InputPath` and `OutputPath` are user-defined directories, make
+sure they exist and you have right permissions. The variable name e.g.
+`SoilPropertyPath` should not be changed. Also, note a `/` is required at
+the end of each line.
As explained above, the "InputPath" directory of the model is considered as
the working/running directory and should include some data required by the
@@ -64,91 +71,3 @@ input_data.xlsx` to the working/running directory. For example:
` cp -r
/projects/0/einf2480/model_parameters/vegetation_property/*
/scratch-shared/EcoExtreML/STEMMUS_SCOPE/input/ `
-
-### Workflow of STEMMUS_SCOPE on Snellius:
-
-This is the same as the workflow of STEMMUS_SCOPE on crib, see section
-[Workflow of STEMMUS_SCOPE on CRIB](./STEMMUS_SCOPE_on_CRIB.md#workflow-of-stemmus_scope-on-crib).
-
-### How to run STEMMUS_SCOPE on Snellius:
-
-1. Log in to Snellius.
-2. Download the source code from this repository or get it using `git clone` in
- a terminal:
-
- ` git clone https://github.com/EcoExtreML/STEMMUS_SCOPE.git `
-
- All the codes can be found in the folder `src` whereas the executable file in
- the folder `exe`.
-
-3. Check `config_file_snellius.txt` and change the paths if needed,
- specifically "InputPath" and "OutputPath".
-4. Follow one of the instructions below:
-
-#### Run using MATLAB that requires a license
-
-In order to use MATLAB, you need to send a request to add you to the user pool
-on Snellius. Then, open the file
-[config_file_snellius.txt](../config_file_snellius.txt) and set the paths. Then,
-run the main script `STEMMUS_SCOPE_exe.m` using MATLAB command line in a terminal on
-a **compute node**:
-
-```bash
-module load 2021
-module load MATLAB/2021a-upd3
-matlab -nodisplay -nosplash -nodesktop -r "STEMMUS_SCOPE_exe('config_file_snellius.txt');exit;"
-```
-
-> To run the codes above on a compute node, you can create a bash script as:
-
-```bash
-#!/bin/bash
-# SLURM settings
-#SBATCH -J stemmus_scope
-#SBATCH -t 01:00:00
-#SBATCH -N 1
-#SBATCH --ntasks=1
-#SBATCH --cpus-per-task=32
-#SBATCH -p thin
-#SBATCH --output=./slurm_%j.out
-#SBATCH --error=./slurm_%j.out
-
-module load 2021
-module load MATLAB/2021a-upd3
-matlab -nodisplay -nosplash -nodesktop -r "STEMMUS_SCOPE_exe('config_file_snellius.txt');exit;"
-```
-
-#### Run using MATLAB Compiler that does not require a license
-
-If you want to run the model as a standalone application, you need MATLAB
-Runtime version `2023a`. This is available on Snellius. You can run the
-model by passing the path of the config file in a terminal on a **compute
-node**:
-
-```bash
-module load 2021
-module load MCR/R2021a.3
-./STEMMUS_SCOPE/exe/STEMMUS_SCOPE config_file_snellius.txt
-```
-
-The bash script `run_stemmus_scope_snellius.sh` in this repository, runs the
-model at 170 sites (default) on a **compute node**. The scripts loops over
-forcing files in the "ForcingPath", creates `sitename_timestamped` working
-directories under "InputPath" directory and copies required data to those
-working dirs. To change the number of sites, open the script and on the last
-line change the parameter `{1..170}`. For example `env_parallel -n1 -j32
---joblog $log_file loop_func ::: {1..170}` will run the model at 32 sites
-simultaneously. For testing purposes, the time of the bash script is set to
-`00:10:00`. Note that the model run can take long for some of the sites. As the
-maxium time wall is 5 days on a partition thin, time can be set to`5-00:00:00`
-for a complete run of the model.
-
- You can run the script in a terminal:
-
-```shell
-cd STEMMUS_SCOPE
-mkdir -p slurm
-sbatch run_stemmus_scope_snellius.sh
-```
-
-This creates a log file per each forcing file in the folder `slurm`.
diff --git a/docs/STEMMUS_SCOPE_on_local_device.md b/docs/STEMMUS_SCOPE_on_local_device.md
deleted file mode 100644
index 8c314bad..00000000
--- a/docs/STEMMUS_SCOPE_on_local_device.md
+++ /dev/null
@@ -1,31 +0,0 @@
-## STEMMUS_SCOPE on local device
-
-You can run STEMMUS_SCOPE locally. Follow the steps below and use `config_file_crib.txt` to configure the paths correctly.
-
-### How to run STEMMUS_SCOPE on local device:
-
-1. Download the source code from this repository or get it using `git clone` in
- a terminal:
-
- ` git clone https://github.com/EcoExtreML/STEMMUS_SCOPE.git `
-
- All the codes can be found in the folder `src` whereas the executable file in
- the folder `exe`.
-
-2. Check `config_file_crib.txt` and change the paths if needed, specifically
- "InputPath" and "OutputPath".
-3. Follow the instructions below:
-
-#### Run using MATLAB Compiler that does not require a license
-
-If you want to run the model as a standalone application, you need MATLAB
-Runtime version `2023a`. To download and install the MATLAB Runtime, follow
-this
-[instruction](https://nl.mathworks.com/products/compiler/matlab-runtime.html).
-The "STEMMUS_SCOPE" executable file is in `STEMMUS_SCOPE/exe` directory
-in this repository. You can run the model by passing the path of the config
-file in a terminal:
-
-```bash
-exe/STEMMUS_SCOPE config_file_crib.txt
-```
\ No newline at end of file
diff --git a/docs/assets/ecoextreml_logo.png b/docs/assets/ecoextreml_logo.png
new file mode 100644
index 00000000..dd892c52
Binary files /dev/null and b/docs/assets/ecoextreml_logo.png differ
diff --git a/docs/getting_started.md b/docs/getting_started.md
new file mode 100644
index 00000000..ac14bd09
--- /dev/null
+++ b/docs/getting_started.md
@@ -0,0 +1,96 @@
+# Requiremnets
+
+## Computig resource
+
+To run the STEMMUS-SCOPE model, you can use one of the following computing resources:
+
+- [CRIB](https://crib.utwente.nl/) is the ITC Geospatial Computing Platform.
+- [Snellius](https://servicedesk.surfsara.nl/wiki/display/WIKI/Snellius) is the
+Dutch National supercomputer hosted at SURF.
+
+Otherwise, you can run the model on your local device if you have the correct
+set of software and data.
+
+## Software
+
+To run the STEMMUS-SCOPE model, you need **one** of the following:
+
+- [MATLAB](https://nl.mathworks.com/products/matlab.html)
+- [MATLAB Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html) on a Unix-like system
+- [Octave](https://octave.org/)
+- [Docker](https://www.docker.com/)
+
+## Model source code
+
+The source code of STEMMUS_SCOPE can be found in the GitHub repository
+[https://github.com/EcoExtreML/STEMMUS_SCOPE](https://github.com/EcoExtreML/STEMMUS_SCOPE)
+under the folder `src`. Download the [latest version of the
+model](https://github.com/EcoExtreML/STEMMUS_SCOPE/releases) from the repository
+or get it using `git clone` in a terminal:
+
+` git clone https://github.com/EcoExtreML/STEMMUS_SCOPE.git `
+
+
+## Data
+
+To run the STEMMUS-SCOPE model, you need to have input data either from in-situ
+measurements or from remote sensing. Before running the model, you need to
+prepare input data and a configuration file for **one site/location**. This can be done using
+[setup()](https://pystemmusscope.readthedocs.io/en/latest/reference/#PyStemmusScope.stemmus_scope.StemmusScope.setup) function
+in the python package [PyStemmusScope](https://pystemmusscope.readthedocs.io/en/latest/). See example datasets below:
+
+=== "Example dataset on Zenodo"
+ A pre-processed example dataset for one site can be found on Zenodo
+ [here](https://zenodo.org/records/10566827).
+
+=== "Data on CRIB"
+ [CRIB](https://crib.utwente.nl/) is the ITC Geospatial Computing Platform.
+
+ {% include-markdown "./STEMMUS_SCOPE_on_CRIB.md" start="### Dataflow of STEMMUS_SCOPE on CRIB" end="### Configuration file" heading-offset=2%}
+
+=== "Data on Snellius"
+ [Snellius](https://servicedesk.surfsara.nl/wiki/display/WIKI/Snellius) is the
+ Dutch National supercomputer hosted at SURF.
+
+ {% include-markdown "./STEMMUS_SCOPE_on_Snellius.md" start="### Dataflow of STEMMUS_SCOPE on Snellius" end="### Configuration file" heading-offset=2%}
+
+## Configuration file
+
+The configuration file is a text file that sets the paths required by the model.
+The configuration file should contain the following information:
+
+```text
+WorkDir=/path_to_working_directory/
+SoilPropertyPath=/path_to_soil_property_data/
+ForcingPath=/path_to_forcing_data/
+Location=AU-DaS
+directional=/path_to_directional_data/
+fluspect_parameters=/path_to_fluspect_parameters_data/
+leafangles=/path_to_leafangles_data/
+radiationdata=/path_to_radiation_data/
+soil_spectrum=/path_to_soil_spectra_data/
+InitialConditionPath=/path_to_soil_initial_condition_data/
+input_data=/path_to_input_data.xlsx_file/
+StartTime=2001-01-01T00:00
+EndTime=2001-01-02T00:00
+InputPath=/path_to_model_input_folder/
+OutputPath=/path_to_model_output_folder/
+```
+
+See example configuration files below:
+
+=== "Example configuration file"
+ An example configuration file can be found
+ [here](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/config_file_template.txt). If
+ [setup()](https://pystemmusscope.readthedocs.io/en/latest/reference/#PyStemmusScope.stemmus_scope.StemmusScope.setup)
+ function in the python package
+ [PyStemmusScope](https://pystemmusscope.readthedocs.io/en/latest/) is used
+ to prepare data, the model configuration file including `InputPath` and
+ `OutputPath` and the data of **one site/location** will be generated
+ automatically.
+
+=== "Example configuration file on CRIB"
+ {% include-markdown "./STEMMUS_SCOPE_on_CRIB.md" start="### Configuration file" heading-offset=2%}
+
+=== "Example configuration file on Snellius"
+ {% include-markdown "./STEMMUS_SCOPE_on_Snellius.md" start="### Configuration file" heading-offset=2%}
diff --git a/docs/index.md b/docs/index.md
new file mode 100644
index 00000000..d14a2278
--- /dev/null
+++ b/docs/index.md
@@ -0,0 +1,7 @@
+# STEMMUS_SCOPE
+
+Integrated code of SCOPE and STEMMUS.
+
+SCOPE is a radiative transfer and energy balance model, and STEMMUS model is a two-phase mass and heat transfer model. For more information about the coupling between these two models, please check [this reference](https://gmd.copernicus.org/articles/14/1379/2021/).
+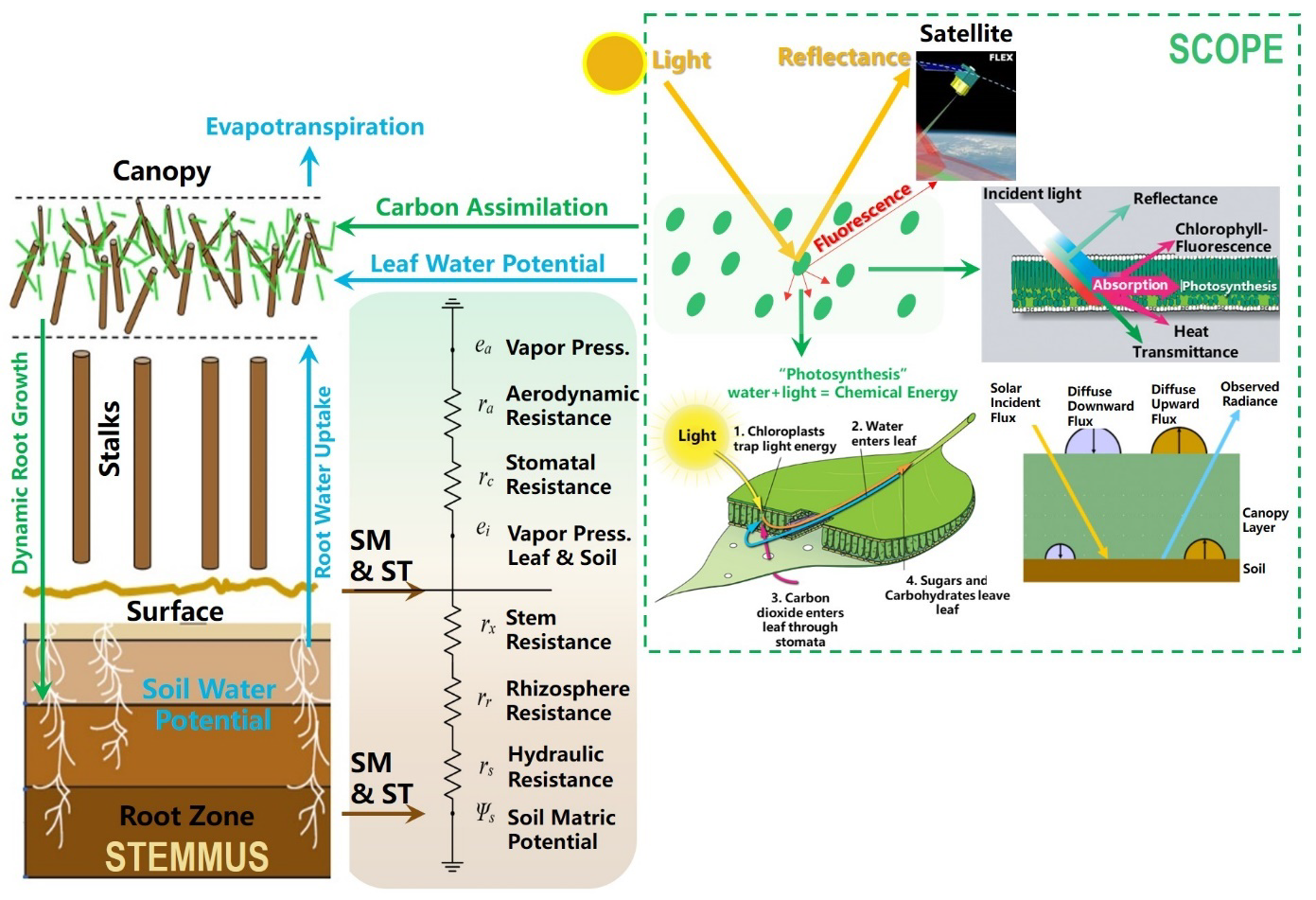
+(by Zeng & Su, 2021)
\ No newline at end of file
diff --git a/docs/requirements.txt b/docs/requirements.txt
new file mode 100644
index 00000000..6253c1f1
--- /dev/null
+++ b/docs/requirements.txt
@@ -0,0 +1,4 @@
+mkdocs
+mkdocs-material
+mkdocs-gen-files
+mkdocs-include-markdown-plugin
\ No newline at end of file
diff --git a/docs/run_model.md b/docs/run_model.md
new file mode 100644
index 00000000..b713cae5
--- /dev/null
+++ b/docs/run_model.md
@@ -0,0 +1,219 @@
+#
+
+> NOTE: The instructions below are meant for users who want to run the model. If
+you want to add changes to the model, see the documentation on ["**Contributing
+guide**"](./CONTRIBUTING.md).
+
+## Workflow of STEMMUS_SCOPE
+
+1. The model reads the forcing file associated with the specified site/location,
+ e.g., `FI-Hyy_1996-2014_FLUXNET2015_Met.nc` from "ForcingPath" and extracts
+ forcing variables in `.dat` format. The `.dat` files are stored in the
+ `InputPath` directory. In addition, the model reads the site information i.e.
+ the location and vegetation parameters.
+2. The model runs step by step until the whole simulation period is completed
+ i.e till the last time step of the forcing data.
+3. The results are saved as binary files temporarily. Then, the binary files are
+ converted to `.csv` files and stored in a `sitename_timestamped` output
+ directory under `OutputPath`.
+
+
+## Run the model with MATLAB
+
+=== "Local device"
+ If you have [MATLAB](https://nl.mathworks.com/products/matlab.html) installed on
+ your device, you can run the model by passing config file path to the
+ variable `CFG` in the main script `STEMMUS_SCOPE.m`.
+
+ As an alternative, you can run the main script using MATLAB command line in a
+ terminal:
+
+ ```bash
+ cd src/
+ matlab -nodisplay -nosplash -nodesktop -r "run('STEMMUS_SCOPE.m');exit;"
+ ```
+=== "CRIB"
+ To open MATLAB desktop on CRIB, click on the `Remote Desktop` in the
+ Launcher. Click on the `Applications`. You will find the 'MATLAB' software
+ under the `Research`. After clicking on 'MATLAB', it asks for your account
+ information that is connected to a MATLAB license. Then, you can run the
+ model by passing config file path to the variable `CFG` in the main script
+ `STEMMUS_SCOPE.m`
+
+=== "Snellius"
+ In order to use MATLAB, you need to send a request to add you to the user pool
+ on Snellius. Then, pass config file path to the variable `CFG` in the main script
+ `STEMMUS_SCOPE.m`. To run the main script `STEMMUS_SCOPE.m` using MATLAB command line in a terminal on
+ a **compute node**:
+
+ ```bash
+ module load 2023
+ module load MATLAB/2023a-upd4
+ cd src/
+ matlab -nodisplay -nosplash -nodesktop -r "run('STEMMUS_SCOPE.m');exit;"
+ ```
+
+ To submit a job to a compute node, see instructions [here](https://servicedesk.surf.nl/wiki/display/WIKI/Example+job+scripts).
+
+## Run the model with MATLAB Runtime
+
+=== "Local device"
+ If you want to run the model as a standalone application, you need [MATLAB
+ Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html)
+ version `2023a`. You don't need a license for MATLAB Runtime. Note that the
+ version of the MATLAB Runtime is tied to the version of MATLAB. The
+ executable file works on the operating system on which the file is created.
+ The file `STEMMUS_SCOPE` under `exe` directory is an executable file of
+ STEMMUS_SCOPE that is created using MATLAB version `2023a` in a **Linux
+ system**.
+
+ Make sure `LD_LIBRARY_PATH` is set correctly, in a terminal:
+
+ ```bash
+ export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/usr/local/MATLAB/MATLAB_Runtime/v2023a/runtime/glnxa64:/usr/local/MATLAB/MATLAB_Runtime/v2023a/bin/glnxa64:/usr/local/MATLAB/MATLAB_Runtime/v2023a/sys/os/glnxa64:/usr/local/MATLAB/MATLAB_Runtime/v2023a/sys/opengl/lib/glnxa64
+ ```
+
+ Change `/usr/local/MATLAB/MATLAB_Runtime/v2023a` to the path where MATLAB Runtime is installed.
+
+ Then, you can run the model in a terminal:
+
+ ```bash
+ ./STEMMUS_SCOPE/exe/STEMMUS_SCOPE config_file_template.txt
+ ```
+
+=== "CRIB"
+ On CRIB, [MATLAB
+ Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html) is
+ not available. Contact the system administrator for more information.
+
+=== "Snellius"
+ [MATLAB
+ Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html) is available on Snellius. You can run the
+ model by passing the path of the config file in a terminal on a **compute
+ node**:
+
+ ```bash
+ module load 2023
+ module load MATLAB/2023a-upd4
+ ./STEMMUS_SCOPE/exe/STEMMUS_SCOPE config_file_snellius.txt
+ ```
+
+ Note that you don't need to set `LD_LIBRARY_PATH` on Snellius. To submit a job to a compute node, see instructions [here](https://servicedesk.surf.nl/wiki/display/WIKI/Example+job+scripts).
+
+=== "BMI interface"
+ You can run the model using the [BMI](https://bmi.readthedocs.io/en/stable/)
+ interface. The BMI interface is available in the script
+ `STEMMUS_SCOPE_exe.m`. For that, you need [MATLAB
+ Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html)
+ version `2023a`. You don't need a license for MATLAB Runtime. Note that the
+ version of the MATLAB Runtime is tied to the version of MATLAB. The
+ executable file works on the operating system on which the file is created.
+ The file `STEMMUS_SCOPE` under `exe` directory is an executable file of
+ STEMMUS_SCOPE that is created using MATLAB version `2023a` in a **Linux
+ system**.
+
+ Make sure `LD_LIBRARY_PATH` is set correctly, in a terminal:
+
+ ```bash
+ export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/usr/local/MATLAB/MATLAB_Runtime/v2023a/runtime/glnxa64:/usr/local/MATLAB/MATLAB_Runtime/v2023a/bin/glnxa64:/usr/local/MATLAB/MATLAB_Runtime/v2023a/sys/os/glnxa64:/usr/local/MATLAB/MATLAB_Runtime/v2023a/sys/opengl/lib/glnxa64
+ ```
+
+ Change `/usr/local/MATLAB/MATLAB_Runtime/v2023a` to the path where MATLAB Runtime is installed.
+
+ To enter the BMI mode:
+
+ ```bash
+ ./STEMMUS_SCOPE/exe/STEMMUS_SCOPE "" bmi
+ ```
+
+ Now, BMI asks you to select one of the steps "initialize", "update" or "finalize".
+ To initialize the model using a config file, type:
+
+ ```bash
+ initialize "path/to/config_file_template.txt"
+ ```
+
+ Change `path/to/config_file_template.txt` to the actual path of the config file.
+
+## Run the model with Octave
+
+=== "Local device"
+ {% include-markdown "./Octave_instructions.md" start="# Running STEMMUS-SCOPE in Octave" end="## VS Code setup" heading-offset=2%}
+
+=== "CRIB"
+ On CRIB, you can use [Octave](https://octave.org/). Allmost all
+ funcationalities of STEMMUS_SCOPE are compatible with Octave, but the
+ execution time is longer than MATLAB. After Octave installation, launch
+ octave and install the following Octave packages:
+
+ ```bash
+ pkg install "https://downloads.sourceforge.net/project/octave/Octave%20Forge%20Packages/Individual%20Package%20Releases/io-2.6.4.tar.gz"
+ pkg install "https://downloads.sourceforge.net/project/octave/Octave%20Forge%20Packages/Individual%20Package%20Releases/statistics-1.4.3.tar.gz"
+ ```
+
+ Then, pass config file path to the variable `CFG` in the main script
+ `STEMMUS_SCOPE.m`. Run the model in a terminal:
+
+ ```bash
+ octave.bat --no-gui --interactive --silent --eval "STEMMUS_SCOPE"
+ ```
+
+=== "Snellius"
+ Since Octave is very slow, it is not recommended to run the model on
+ Snellius using Octave. Use MATLAB Runtime instead.
+
+## Run the model with Docker
+
+=== "Local device"
+ If you have [Docker](https://www.docker.com/) installed on your device, you
+ can run the model using the docker image `ecoextreml/stemmus_scope` and
+ [BMI](https://bmi.readthedocs.io/en/stable/) interface. The docker image is available
+ on
+ [EcoExtreML](https://github.com/EcoExtreML/STEMMUS_SCOPE/pkgs/container/stemmus_scope).
+ You can pull the `latest` image using the following command:
+
+ ```bash
+ docker pull ghcr.io/ecoextreml/stemmus_scope:latest
+ ```
+
+ Then, mount the directories and run the model in a terminal:
+
+ ```bash
+ docker run -u $(id -u):$(id -g) -v /path_to_input_folder:/path_to_input_folder -v /path_to_output_folder:/path_to_output_folder -it ghcr.io/ecoextreml/stemmus_scope:latest
+ ```
+
+ With `-it` option, you can enter the docker interactive mode.
+ Now, BMI asks you to select one of the steps "initialize", "update" or "finalize".
+ To initialize the model using a config file, type:
+
+ ```bash
+ initialize "path/to/config_file_template.txt"
+ ```
+
+ Make sure that the input and output directories are mounted correctly
+ and `config_file_template.txt` is available in the input directory.
+ Also, remove the container after running the model:
+
+ ```bash
+ docker rm container_id
+ ```
+
+=== "CRIB"
+ If [Docker](https://www.docker.com/) is installed on CRIB, follow the same
+ instructions as for the `Local device`.
+
+=== "Snellius"
+ [Docker](https://www.docker.com/) option is not available on Snellius. You
+ need to use
+ [Apptainer](https://apptainer.org/docs/user/main/introduction.html), see
+ instructions
+ [here](https://servicedesk.surf.nl/wiki/pages/viewpage.action?pageId=30660251).
+
+## Example workflow of running the model
+
+1. Download the [latest version of the
+ model](https://github.com/EcoExtreML/STEMMUS_SCOPE/releases).
+2. Download the example dataset from Zenodo
+ [here](https://zenodo.org/records/10566827) that includes the configuration
+ file.
+3. Choose one of the options above to run the model.
diff --git a/mkdocs.yml b/mkdocs.yml
new file mode 100644
index 00000000..88e7f838
--- /dev/null
+++ b/mkdocs.yml
@@ -0,0 +1,55 @@
+site_name: STEMMUS_SCOPE documentation
+repo_url: https://github.com/EcoExtreML/STEMMUS_SCOPE
+repo_name: STEMMUS_SCOPE
+
+nav:
+ - Introduction: index.md
+ - Getting started: getting_started.md
+ - Running the model: run_model.md
+ - Contributing guide: CONTRIBUTING.md
+
+theme:
+ name: material
+ custom_dir: docs/assets
+ logo: assets/ecoextreml_logo.png
+ features:
+ - navigation.instant
+ - navigation.tabs
+ - navigation.tabs.sticky
+ icon:
+ repo: fontawesome/brands/github-alt
+
+ palette:
+ # Palette toggle for light mode
+ - scheme: default
+ toggle:
+ icon: material/weather-sunny
+ name: Switch to dark mode
+ primary: light green
+ accent: green
+
+ # Palette toggle for dark mode
+ - scheme: slate
+ toggle:
+ icon: material/weather-night
+ name: Switch to light mode
+ primary: blue grey
+ accent: teal
+
+plugins:
+- include-markdown
+- search
+- autorefs
+
+markdown_extensions:
+ - pymdownx.highlight:
+ anchor_linenums: true
+ - pymdownx.superfences
+ - pymdownx.details
+ - admonition
+ - pymdownx.tabbed:
+ alternate_style: true
+ - attr_list
+
+extra:
+ generator: false
diff --git a/pull_request_template.md b/pull_request_template.md
index be1b31af..de2db406 100644
--- a/pull_request_template.md
+++ b/pull_request_template.md
@@ -6,8 +6,10 @@ Please add a description of the changes proposed in the pull request.
- [ ] @mentions of the person or team responsible for reviewing proposed changes.
- [ ] This pull request has a descriptive title.
- [ ] Code is written according to the [guidelines](http://cnl.sogang.ac.kr/cnlab/lectures/programming/matlab/Richard_Johnson-MatlabStyle2_book.pdf).
-- [ ] The checks by [MISS_HIT style checker and linter](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/CONTRIBUTING.md#follow-matlab-style-guidelines), below the pull request, are successful (green).
+- [ ] The checks by [MISS_HIT style checker and
+ linter](./docs/CONTRIBUTING.md#miss_hit-checks), below the pull request, are
+ successful (green).
- [ ] Documentation is available.
- [ ] Add changes to the [changelog file](CHANGELOG.md) under section `Unreleased`.
-- [ ] Model runs successfully.
+- [ ] Model runs successfully, see [tests](./docs/CONTRIBUTING.md#testing-new-changes).
- [ ] Ask a meinatainer to re-generate exe file if matlab codes are changed. About how to create an exe file, see [exe readme](https://github.com/EcoExtreML/STEMMUS_SCOPE/blob/main/run_model_on_snellius/exe/README.md).
diff --git a/run_model_on_snellius/README.md b/run_model_on_snellius/README.md
index 92cc0bd6..bd12273b 100644
--- a/run_model_on_snellius/README.md
+++ b/run_model_on_snellius/README.md
@@ -7,3 +7,25 @@ This folder contains files that are needed for running `STEMMUS_SCOPE` on Snelli
- `config_file_snellius.txt` is the model config file
Before submitting the job via `sbatch run_stemmus_scope_snellius.sh`, make sure that conda environment `pystemmusscope` is created, see the [environment file](https://github.com/EcoExtreML/STEMMUS_SCOPE_Processing/blob/main/environment.yml). Also, set the `WorkDir` and `NumberOfTimeSteps` in the model config file e.g. `config_file_snellius.txt`.
+
+The bash script `run_stemmus_scope_snellius.sh` in this repository, runs the
+model at 170 sites (default) on a **compute node**. The scripts loops over
+forcing files in the "ForcingPath", creates `sitename_timestamped` working
+directories under "InputPath" directory and copies required data to those
+working dirs. To change the number of sites, open the script and on the last
+line change the parameter `{1..170}`. For example `env_parallel -n1 -j32
+--joblog $log_file loop_func ::: {1..170}` will run the model at 32 sites
+simultaneously. For testing purposes, the time of the bash script is set to
+`00:10:00`. Note that the model run can take long for some of the sites. As the
+maxium time wall is 5 days on a partition thin, time can be set to`5-00:00:00`
+for a complete run of the model.
+
+You can run the script in a terminal:
+
+```shell
+cd STEMMUS_SCOPE
+mkdir -p slurm
+sbatch run_stemmus_scope_snellius.sh
+```
+
+This creates a log file per each forcing file in the folder `slurm`.
diff --git a/run_model_on_snellius/exe/README.md b/run_model_on_snellius/exe/README.md
index 4d558c63..eb40f7f7 100644
--- a/run_model_on_snellius/exe/README.md
+++ b/run_model_on_snellius/exe/README.md
@@ -1,14 +1,12 @@
# Create an executable file of STEMMUS_SCOPE
If you want to run the STEMMUS_SCOPE model, you need to run it using MATLAB or
-[MATLAB Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html). You don't need a license for MATLAB Runtime.
+[MATLAB Runtime](https://nl.mathworks.com/products/compiler/matlab-runtime.html). You don't need a license for MATLAB Runtime.
The file `STEMMUS_SCOPE` under `exe` directory is an executable file of
STEMMUS_SCOPE that is created using MATLAB version `2023a` in a Linux system.
Note that the version of the MATLAB Runtime is tied to the version of MATLAB.
-For more information, see [How to run STEMMUS_SCOPE on
-CRIB](../README.md#how-to-run-stemmus_scope-on-crib) or [How to run
-STEMMUS_SCOPE on Snellius](../README.md#how-to-run-stemmus_scope-on-snellius).
+For more information, see documentation.
To re-create an executable file of `STEMMUS_SCOPE`, you need a license for [MATLAB compiler](https://nl.mathworks.com/products/compiler.html).
In order to use MATLAB on Snellius, you need to send a request to add you to the