Back to Projects List
- Maximilian Fischer (German Cancer Research Center, Germany)
- Andrey Fedorov (Brigham and Women’s Hospital, USA)
- Marco Nolden (German Cancer Research Center, Germany)
- Philipp Schader (German Cancer Research Center, Germany)
- David Clunie (PixelMed Publishing, USA)
- Daniela Schacherer (Fraunhofer MEVIS, Germany)
- André Homeyer (Fraunhofer MEVIS, Germany)
- Curtis Lisle (KnowledgeVis, USA)
- Theodore Aptekarev (Freelancer)
- Davide Punzo (Freelancer)
- Igor Octaviano (Radical Imaging)
Project channel on Discord: #dicom-wsi
The Imaging Data Commons (IDC) portal is a cloud based repository of public cancer imaging data, and inclludes access to histopathological image data in the DICOM standard. In recent years increasingly more automated image analysis algorithms for pathology data emerged. However they are mainly developed for proprietary WSI vendor file formats and not for the standardized DICOM WSI file format as input. In this project a tumor classification algorithm shall be developed for DICOM WSI files as input file format. The aim of this project is to develop an end to end Google Colab notebook to define a data cohort from the IDC database. The selected cohort is retrieved in the DICOM WSI standard and an examplary deep learning based image analysis algorithm is applied on the selected DICOM studies. As a use case task we select a classification algorithm, which is a common downstream task in computational pathology. The algorithm generates for the selected DICOM WSI files a tumor heatmap and provides the analysis result as DICOM parametric map, which can be visualized together with WSI image in the interoperable web-based slide microscopy viewer and annotation tool (SLIM), which is fully integrated in the IDC database. A basis of this project provides the DICOM WSI support in the Kaapana platform, which also shall be improved in this project. The custom DICOM conversion pipeline for WSI files, which is integrated in the platform shall be improved and further applications with DICOM WSI data integrated in the platform.
- An analysis pipeline to visualize the analysis results for IDC digital pathology data
- Deploy a deep learning based tumor classification algorithm via Google Colab
- Develop further pathology applications in the Kaapana platform
- Define Query/Retrieve operations to define a data cohort from IDC portal
- Test several image analysis algorithms for tumor classification
- Store the result in a DICOM WSI file format
- Integrate workflow in Google Colab
- Had a project kick-off meeting to discuss the plan.
- Agreed for Max and Andrey will work to set up initial part of the colab notebook that searches and downloads WSI from IDC and extracts tiles, then this can be used both by Max and Curt for workflow development
- Agreed to use Colab notebook to set up conversion pipeline using David and google converter
- Established a notebook here to document/share recipes for using various conversion tools and validation of the resulting images. Andrey will continue developing this after the PW.
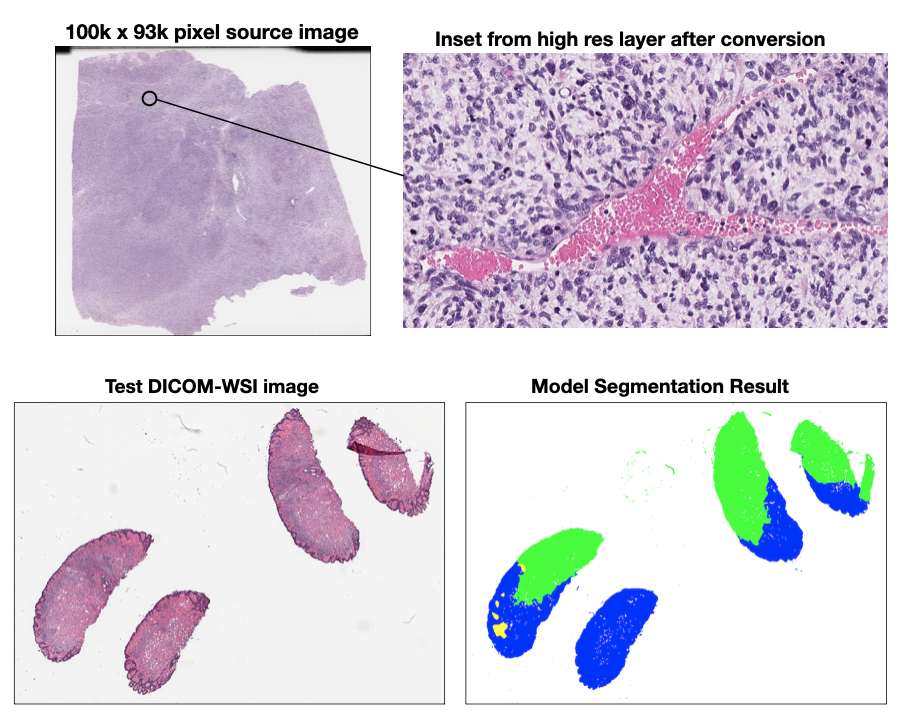
- Curt and David tested three DICOM-WSI converters (PixelMed's converter, Google's wsi2dcm, and the wsidicomizer) using a pyramidal Aperio cancer image from NCI. All converters were able to run on the image, but produced varying results. PixelMed produced the most complete set of layers and DICOM tags with wsidicomizer second and Google's converter coming in third place. (See the high-res example inset below)
- We made good progress adapting a pretrained tissue segmentation algorithm developed by NCI to run on a converted DICOM-WSI source image instead of the proprietary original format the model was designed for. (See below for the sample CMU-1 DICOM WSI and the segmentation output generated from the model.) This effort will contue after the project week. Some WSIs are compressed with JPEG2000, which can present decompression problems in some conversion packages.
- Made progress developing a Colab notebook "recipe" for deploying Slim viewer on GCP via Firebase. This is not yet completed, but made great progress. Notebook is here, Andrey will continue working on this following the PW.
- worfklow used by IDC to create DICOM WSI: https://github.com/ImagingDataCommons/idc-wsi-conversion
- Colab notebooks with examples of using DICOM WSI in analysis workflows: https://github.com/ImagingDataCommons/IDC-Examples/tree/master/notebooks/pathomics
- DICOM WSI converter from Google: https://github.com/GoogleCloudPlatform/wsi-to-dicom-converter
- sample image https://cytomine.com/collection/cmu-1/cmu-1-svs
- Dropbox folder for storing related artifacts
- Max github repo: https://github.com/maxfscher/DICOMwsiWorkflow
- Colab Notebook for the conversion process https://colab.research.google.com/drive/1sbuGggwmbE-JgkO8LS5ndXIzLpf7yhKd?usp=sharing
- Slim deployment instructions (WIP): https://docs.google.com/document/d/1r6r8w4FZnzeQO47TDn9DTj78nTjDsxuy4jSULmrCTwM/edit
- wsidicomizer repository: https://github.com/imi-bigpicture/wsidicomizer