In this tutorial, we will go through a scaling exercise to link the field spectral reflectance and airborne spectrometer orthorectified surface directional reflectance. We will also tie in the plant foliar traits data, which contains the geographic information and other metadata about the plants measured for associated with the field spectra data.
-Field spectral reflectance provide information about individual leaves or foliage, and samples such as these are often used as spectral end-members in classification applications. A collection of these spectral data can comprise a spectral library, eg. the USGS Spectral Library. This NEON data product is collected on an opportunistic basis, typically in conjunction with NEON Airborne Observation Platform (AOP) overflights and in coordination with TOS Canopy Foliage Sampling. AOP typically collects spectra at 1-2 sites per year, so it is currently available at a subset of the NEON sites. The tutorial also demonstrates how to programmatically determine which data are available.
+In this tutorial, we will demonstrate a scaling exercise to link the NEON field spectral reflectance and airborne spectrometer orthorectified surface directional reflectance datasets. We will also tie in the plant foliar traits data, which contains the geographic information and other metadata about the plants associated with the field spectra data.
+Field spectral reflectance provide information about individual leaves or foliage, and samples such as these are often used as spectral end-members in classification applications. A collection of these spectral data can comprise a spectral library, eg. the USGS Spectral Library. This NEON data product is collected on an opportunistic basis, typically in conjunction with NEON Airborne Observation Platform (AOP) overflights and in coordination with Terrestrial Observation System (TOS) Canopy Foliage Sampling. AOP typically collects spectra at 1-2 sites per year, so it is currently available at a subset of the NEON sites. The tutorial also demonstrates how to programmatically determine which data are available.
Learning Objectives
After completing this activity, you will be able to:
@@ -313,7 +313,7 @@Foliar Trait Data
#View(foliar_trait_info$siteCodes$availableDataUrls) -We can get the availableDataUrls of the RMNP site as follows:
We can get the availableDataUrls of the RMNP site as follows. This step is not required, but as we showed with the field spectral data, this is a way to see what foliar trait data are available for a given site, since the canopy foliar sampling does not occur every year.
# view the RMNP foliar trait available data urls
foliar_trait_info$siteCodes[which(foliar_trait_info$siteCodes$siteCode == 'RMNP'),c("availableDataUrls")]
@@ -341,7 +341,7 @@ Foliar Trait Data
Since the spectra_top_black table includes the wavelength and reflectance values for all 426 bands, we can first subset it to only include a single wavelength.
spectra_traits <- merge(spectra_top_black,foliar_traits_loc,by="sampleID")
-# display values of only first wavelength
+# display values of only first wavelength for each sample
spectra_traits_sub <- merge(spectra_top_black[spectra_top_black$wavelength == 350,],foliar_traits_loc,by="sampleID")
@@ -373,8 +373,6 @@ Foliar Trait Data
## 23 FSP_RMNP_20200707_2058 PIPOS 7.4 73.0 456379.8 4450485 RMNP.03868.2020
## 24 FSP_RMNP_20200716_1202 PSME 16.4 338.2 459056.9 4450435 RMNP.03836.2020
## 25 FSP_RMNP_20200713_1117 PICOL 9.3 25.4 460126.2 4449769 RMNP.03626.2020
-
-#head(spectra_traits[c("spectralSampleID","taxonID","stemDistance","stemAzimuth","adjEasting","adjNorthing","crownPolygonID")],1)
Great, now we have the field spectra data and the corresponding locations of the leaf-clip samples.
Airborne Reflectance Data
@@ -448,7 +446,7 @@ Airborne Reflectance Data
return(refl_out) # return the terra raster object
}
Now that we’ve defined these functions, we can run lapply on the band2Raster function, using all the bands. This may take a minute or two to run.
Now that we’ve defined these functions, we can run lapply on the band2Raster function, using all the bands. This may take a minute or two to complete.
# get the relevant metadata using the geth5metadata function
h5_meta <- geth5metadata(h5_file)
@@ -471,7 +469,7 @@ Airborne Reflectance Data
We can also plot an RGB image. First we’ll run lapply on the band2Raster function, this time using a list only consisting of the RGB bands:
rgb <- list(58,34,19)
-# lapply tells R to apply the function to each element in the list
+# lapply applies the function to each element in the RGB list
rgb_list <- lapply(rgb,
FUN = band2Raster,
@@ -490,8 +488,6 @@ Airborne Reflectance Data
tree_loc <- vect(cbind(fsp_rmnp_picol_20200720_1304$adjEasting,
fsp_rmnp_picol_20200720_1304$adjNorthing), crs=h5_meta$crs)
-# plot
-
plot(tree_loc, col="red", add = T)
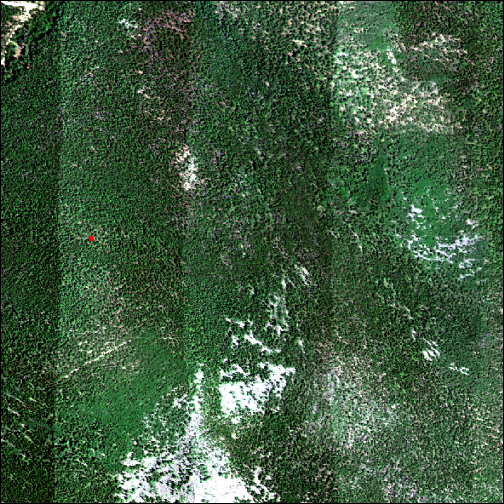
@@ -510,8 +506,6 @@ Airborne Reflectance Data
cbind(fsp_rmnp_picol_20200720_1304$adjEasting[1],
fsp_rmnp_picol_20200720_1304$adjNorthing[1]))
-
-
refl_air_df <- data.frame(t(refl_air))
refl_air_df$wavelengths <- h5_meta$wavelengths
@@ -520,8 +514,6 @@ Airborne Reflectance Data
refl_air_df$reflectance <- refl_air_df$reflectance/10000 #scale by reflectance scale factor (10,000)
-
-
picol_air_plot <- ggplot(refl_air_df, aes(x=wavelength, y=reflectance)) + geom_line() + ylim(0,.25)
print(picol_air_plot + ggtitle("Airborne Reflectance Spectra of PICOL at RMNP"))
@@ -558,7 +550,7 @@ Plotting Leaf-Clip and Ai
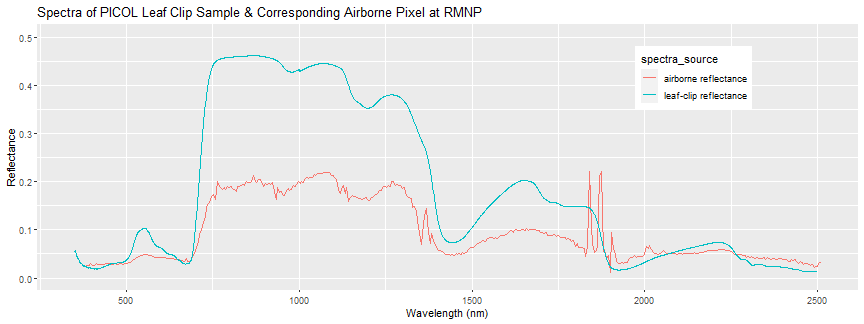
Each reflectance pixel represents a 1m x 1m area, so it contains an average spectra of all the vegetation and non-vegetation that are contained in that area. For example, a single pixel could contain the reflectance of a mix of leaves as well as branches, and any gaps in the tree canopy (eg. the ground under the tree). This averaging is called “spectral mixing” and you can perform “spectral un-mixing” to decompose an average spectra into it’s component parts. These leaf clip spectra can be used as “end-members”, in classification applications, for example.
Also, it is important to understand that there are uncertainties in the ASD measurements as well as the airborne spectra. There may be some geographic uncertainty associated with both the airborne data (which has 0.5 m horizontal resolution), as well as with the field sample geolocations. There is also some uncertainty in the spectral data itself. For example, there is uncertainty resulting from the atmospheric correction applied when generating the reflectance data from the at-sensor-radiance. Weather conditions during the flight are important to consider as well. The Airborne Observation Platform attempts to collect the coincident overflights in clear weather, but this may not always be the case. For more on hyperspectral uncertainties and variation, please refer to the Spectrometer Mosaic ATBD (Algorithm Theoretical Basis Document).
Incorporating the Tree Crown Polygon Shapefiles
@@ -633,29 +625,24 @@Incorporating the Tree
combined_df <- bind_rows(fsp_rmnp_picol_20200720_1304[c("wavelength","reflectance")],picol_crown_df[c("wavelength","reflectance")])
-
-
# add a new column to indicate spectra data source
combined_df$spectra_source <- c(rep("leaf-clip reflectance", nrow(fsp_rmnp_picol_20200720_1304)), rep("airborne reflectance", nrow(picol_crown_df)))
-
-
spectra_crown_plot <- ggplot() +
geom_line(data=combined_df, aes(x=wavelength, y=reflectance, color=spectra_source), show.legend=TRUE) +
labs(x="Wavelength (nm)", y="Reflectance") + theme(legend.position = c(0.8, 0.8)) + ylim(0, 0.5)
-
-
print(spectra_crown_plot + ggtitle("Spectra of PICOL Leaf Clip Sample & Corresponding Airborne Tree-Crown Pixels at RMNP"))
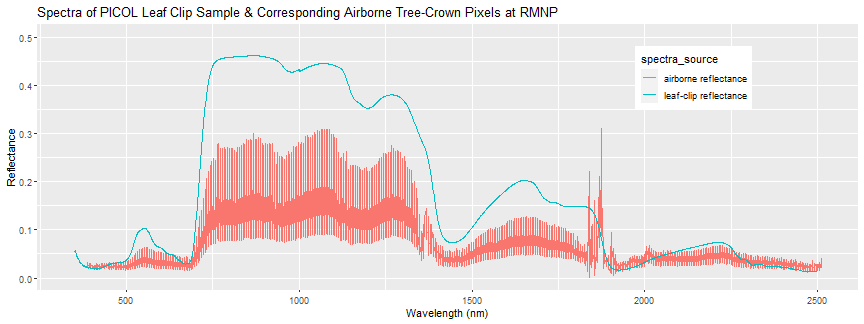
-

Again we can see there is some discrepancy between the airborne and field spectra, and that there is a decent amount of variation in the spectra of all the pixels contained in the tree crown. The leaf-clip spectra is one “end-member” that contributes to the 1-m pixel combined reflectance. This exploratory analysis
-Next Steps
-This tutorial demonstrates a simple example of linking the field spectra data and AOP reflectance data for a single sample, using geographic data included in the foliar sampling datasets, as well as crown polygon shapefile data. This is intended to provide a bouncing-off point for more exploratory analysis and/or in-depth research. What else might you want to explore, using these linked datasets?
+Again we can see there is some discrepancy between the airborne and field spectra, and there is a decent amount of variation between the spectra of the pixels contained within the tree crown polygon. The leaf-clip spectra is one “end-member” that contributes to the 1-m pixel combined reflectance. The exploratory anaysis demonstrated in this tutorial is an example of some steps you might take to get a feel for the data. We recommend you explore some other samples, or other field spectra datasets and build off this R code to answer some questions you come up with as you start digging into the data.
+Next Steps (On Your Own)
+This tutorial demonstrates a simple example of linking the field spectra data and AOP airborne reflectance data for a single sample, using geographic data included in the foliar traits datasets, as well as crown polygon shapefile data. This is intended to provide a bouncing-off point for more exploratory analysis and/or in-depth research. What else might you want to explore, using these linked datasets?
Here are some ideas to pursue on your own:
-
@@ -666,5 +653,6 @@
Next Steps