How to cite COB
+ + + + + + +How to contribute to COB
+ + + + + + +Editors documentation
+-
+
- Do not edit cob.owl directly. Instead edit cob-edit.owl +
- Do not edit cob-to-external.owl directly. Instead edit the tsv and run the Makefile. +
Run a COB release:
+See standard ODK release docs here.
+Run COB integration tests:
+All COB tests:
+cd src/ontology
+sh run.sh make cob_test
+Basic COB tests:
+cd src/ontology
+sh run.sh make main_test
+Run regular ontology QC tests:
+cd src/ontology
+sh run.sh make IMP=false test
+Exploring COB
+Browse
+COB is available on the following browsers:
+ +You can also enter any COB class PURL into a browser to resolve it; e.g. http://purl.obolibrary.org/obo/COB_0000011 (atom)
+Files
+-
+
- End products
-
+
- cob.owl: the current version of COB containing new terms in the COB namespace +
- cob-to-external.owl COB terms and OBO Foundry counterparts with proposed equivalencies +
- cob-examples.owl: additional child terms as examples. This file is edited by hand. +
+ - Editors/Source files +
- cob-edit.owl: the editors version +
- cob-to-external.tsv: TSV source of cob-to-external +
- Makefile: workflow for build COB +
COB demo
+As a demonstration of how COB could be used to unify OBO ontologies in the future we produce an ontology:
+ +This has selected subsets of certain ontologies merged in with +cob-to-external. It is incomplete and messy. Please see the Makefile +for how to add more to it.
+ + + + + + +A brief history of COB
+The following page gives an overview of the history of COB.
+ + + + + + +COB Ontology Documentation
+This project is an attempt to bring together key terms from a wide range of Open Biological and Biomedical Ontology (OBO) projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms. Our plan is to keep this ontology small, but ensure that one or more COB terms can be used as the root of any given OBO library ontology.
+Editors Guide
+You can find descriptions of the standard ontology engineering workflows here.
+Issue Tracker
+If you are responsible for an OBO ontology you can see any tickets that pertain to your ontology by looking for the label with your ontology ID.
+For example:
+ + + + + + + +Integration Tests
+Currently we have two kinds:
+-
+
- Coherency: testing if an ontology is coherent when combined with COB + ext +
- Coverage: testing if an ontology has classes that are not subclasses (direct or indirect) of COB classes +
See the products folder
+ + + + + + +Placing OBO ontologies under COB
+COB is intended as a central place where different ontologies within +OBO can discuss and agree where the different root terms across OBO +belong in the COB hierarchy.
+The source of truth for this is a file "COB to External"
+COB to External
+The source for the file is in the src/ontology/components folder in GitHub:
+ +The file is in SSSOM tsv format.
+Example entries
+The following entries use the owl:equivalentClass predicate to indicate that the COB IDs are equivalent to the the corresponding OBO concept.
+| subject_id | +subject_label | +predicate_id | +object_id | +object_label | +notes | +
|---|---|---|---|---|---|
| COB:0000003 | +mass | +owl:equivalentClass | +PATO:0000125 | +mass | +. | +
| COB:0000004 | +charge | +owl:equivalentClass | +PATO:0002193 | +electric | +. | +
| COB:0000006 | +material entity | +owl:equivalentClass | +BFO:0000040 | +material entity | +. | +
| COB:0000008 | +proton | +owl:equivalentClass | +CHEBI:24636 | +proton | +. | +
| COB:0000017 | +cell | +owl:equivalentClass | +CL:0000000 | +cell | +. | +
| COB:0000026 | +processed material entity | +owl:equivalentClass | +OBI:0000047 | +processed material | +. | +
| COB:0000031 | +immaterial entity | +owl:equivalentClass | +BFO:0000041 | +immaterial entity | +. | +
| COB:0000033 | +realizable | +owl:equivalentClass | +BFO:0000017 | +realizable entity | +. | +
Multiple OBO IDs for one COB ID
+Ideally no COB ID is equivalent to more than one OBO ID, but in +practice different ontologies have minted IDs for what turned out to +be the same concept. COB serves as a useful clearinghouse for making +these lack of orthogonality transparent.
+For example, the COB concept of "organism" (which includes viruses):
+| subject_id | +subject_label | +predicate_id | +object_id | +object_label | +notes | +
|---|---|---|---|---|---|
| COB:0000022 | +organism | +owl:equivalentClass | +NCBITaxon:1 | +root | +. | +
| COB:0000022 | +organism | +owl:equivalentClass | +OBI:0100026 | +organism | +. | +
| COB:0000022 | +organism | +owl:equivalentClass | +CARO:0001010 | +organism or virus or viroid | +. | +
subClass links
+we can also declare the OBO class to be a subclass of the COB class. For example, the concept of a cell in the plant ontology is the species equivalent of the species-generic COB cell, so formally it is a subclass:
+| subject_id | +subject_label | +predicate_id | +object_id | +object_label | +notes | +
|---|---|---|---|---|---|
| COB:0000017 | +cell | +owl:equivalentClass | +CL:0000000 | +cell | +. | +
| COB:0000017 | +cell | +sssom:superClassOf | +PO:0009002 | +plant cell | +. | +
| COB:0000018 | +native cell | +owl:equivalentClass | +CL:0000003 | +native cell | +. | +
| COB:0000018 | +native cell | +sssom:superClassOf | +XAO:0003012 | +xenopus cell | +. | +
| COB:0000018 | +native cell | +sssom:superClassOf | +ZFA:0009000 | +zebrafish cell | +. | +
| COB:0000018 | +native cell | +sssom:superClassOf | +PO:0025606 | +native plant cell | +. | +
| COB:0000019 | +cell in vitro | +owl:equivalentClass | +CL:0001034 | +cell in vitro | +. | +
Similarly, we can record the fact that we are committing to group SO entities as material entities
+| subject_id | +subject_label | +predicate_id | +object_id | +object_label | +notes | +
|---|---|---|---|---|---|
| COB:0000006 | +material entity | +sssom:superClassOf | +SO:0000110 | +sequence_feature | +. | +
| COB:0000006 | +material entity | +sssom:superClassOf | +SO:0001060 | +sequence_variant | +. | +
| COB:0000006 | +material entity | +sssom:superClassOf | +SO:0001260 | +sequence_collection | +. | +
Formal lack of commitment
+We can record a formal lack of commitment in cob-to-external:
+| subject_id | +subject_label | +predicate_id | +object_id | +object_label | +notes | +
|---|---|---|---|---|---|
| COB:0000011 | +atom | +skos:closeMatch | +CHEBI:33250 | +atom | +. | +
| COB:0000013 | +molecular entity | +skos:closeMatch | +CHEBI:23367 | +molecular entity | +No exact match in CHEBI | +
| COB:0000013 | +molecular entity | +skos:closeMatch | +CHEBI:25367 | +molecule | +This is electrically neutral in CHEBI but in COB it is generic. We should make it union of molecule and polyatomic ion | +
We cannot make these equivalent because we would end up with incoherencies in COB
+Proposing changes to cob-to-external
+Anyone is welcome to make Pull Requests on the cob-to-external file. The pull request will be discussed openly and transparently, and anyone can make comments on the GitHub PR
+ + + + + + +Introduction to Continuous Integration Workflows with ODK
+Historically, most repos have been using Travis CI for continuous integration testing and building, but due to +runtime restrictions, we recently switched a lot of our repos to GitHub actions. You can set up your repo with CI by adding +this to your configuration file (src/ontology/cob-odk.yaml):
+ci:
+ - github_actions
+When updateing your repo, you will notice a new file being added: .github/workflows/qc.yml.
This file contains your CI logic, so if you need to change, or add anything, this is the place!
+Alternatively, if your repo is in GitLab instead of GitHub, you can set up your repo with GitLab CI by adding +this to your configuration file (src/ontology/cob-odk.yaml):
+ci:
+ - gitlab-ci
+This will add a file called .gitlab-ci.yml in the root of your repo.
Editors Workflow
+The editors workflow is one of the formal workflows to ensure that the ontology is developed correctly according to ontology engineering principles. There are a few different editors workflows:
+-
+
- Local editing workflow: Editing the ontology in your local environment by hand, using tools such as Protégé, ROBOT templates or DOSDP patterns. +
- Completely automated data pipeline (GitHub Actions) +
- DROID workflow +
This document only covers the first editing workflow, but more will be added in the future
+Local editing workflow
+Workflow requirements:
+-
+
- git +
- github +
- docker +
- editing tool of choice, e.g. Protégé, your favourite text editor, etc +
1. Create issue
+Ensure that there is a ticket on your issue tracker that describes the change you are about to make. While this seems optional, this is a very important part of the social contract of building an ontology - no change to the ontology should be performed without a good ticket, describing the motivation and nature of the intended change.
+2. Update main branch
+In your local environment (e.g. your laptop), make sure you are on the main (prev. master) branch and ensure that you have all the upstream changes, for example:
git checkout master
+git pull
+3. Create feature branch
+Create a new branch. Per convention, we try to use meaningful branch names such as: +- issue23removeprocess (where issue 23 is the related issue on GitHub) +- issue26addcontributor +- release20210101 (for releases)
+On your command line, this looks like this:
+git checkout -b issue23removeprocess
+4. Perform edit
+Using your editor of choice, perform the intended edit. For example:
+Protégé
+-
+
- Open
src/ontology/cob-edit.owlin Protégé
+ - Make the change +
- Save the file +
TextEdit
+-
+
- Open
src/ontology/cob-edit.owlin TextEdit (or Sublime, Atom, Vim, Nano)
+ - Make the change +
- Save the file +
Consider the following when making the edit.
+-
+
- According to our development philosophy, the only places that should be manually edited are:
-
+
src/ontology/cob-edit.owl
+- Any ROBOT templates you chose to use (the TSV files only) +
- Any DOSDP data tables you chose to use (the TSV files, and potentially the associated patterns) +
- components (anything in
src/ontology/components), see here.
+
+ - Imports should not be edited (any edits will be flushed out with the next update). However, refreshing imports is a potentially breaking change - and is discussed elsewhere. +
- Changes should usually be small. Adding or changing 1 term is great. Adding or changing 10 related terms is ok. Adding or changing 100 or more terms at once should be considered very carefully. +
4. Check the Git diff
+This step is very important. Rather than simply trusting your change had the intended effect, we should always use a git diff as a first pass for sanity checking.
+In our experience, having a visual git client like GitHub Desktop or sourcetree is really helpful for this part. In case you prefer the command line:
+git status
+git diff
+5. Quality control
+Now it's time to run your quality control checks. This can either happen locally (5a) or through your continuous integration system (7/5b).
+5a. Local testing
+If you chose to run your test locally:
+sh run.sh make IMP=false test
+This will run the whole set of configured ODK tests on including your change. If you have a complex DOSDP pattern pipeline you may want to add PAT=false to skip the potentially lengthy process of rebuilding the patterns.
sh run.sh make IMP=false PAT=false test
+6. Pull request
+When you are happy with the changes, you commit your changes to your feature branch, push them upstream (to GitHub) and create a pull request. For example:
+git add NAMEOFCHANGEDFILES
+git commit -m "Added biological process term #12"
+git push -u origin issue23removeprocess
+Then you go to your project on GitHub, and create a new pull request from the branch, for example: https://github.com/INCATools/ontology-development-kit/pulls
+There is a lot of great advise on how to write pull requests, but at the very least you should:
+- mention the tickets affected: see #23 to link to a related ticket, or fixes #23 if, by merging this pull request, the ticket is fixed. Tickets in the latter case will be closed automatically by GitHub when the pull request is merged.
+- summarise the changes in a few sentences. Consider the reviewer: what would they want to know right away.
+- If the diff is large, provide instructions on how to review the pull request best (sometimes, there are many changed files, but only one important change).
7/5b. Continuous Integration Testing
+If you didn't run and local quality control checks (see 5a), you should have Continuous Integration (CI) set up, for example: +- Travis +- GitHub Actions
+More on how to set this up here. Once the pull request is created, the CI will automatically trigger. If all is fine, it will show up green, otherwise red.
+8. Community review
+Once all the automatic tests have passed, it is important to put a second set of eyes on the pull request. Ontologies are inherently social - as in that they represent some kind of community consensus on how a domain is organised conceptually. This seems high brow talk, but it is very important that as an ontology editor, you have your work validated by the community you are trying to serve (e.g. your colleagues, other contributors etc.). In our experience, it is hard to get more than one review on a pull request - two is great. You can set up GitHub branch protection to actually require a review before a pull request can be merged! We recommend this.
+This step seems daunting to some hopefully under-resourced ontologies, but we recommend to put this high up on your list of priorities - train a colleague, reach out!
+9. Merge and cleanup
+When the QC is green and the reviews are in (approvals), it is time to merge the pull request. After the pull request is merged, remember to delete the branch as well (this option will show up as a big button right after you have merged the pull request). If you have not done so, close all the associated tickets fixed by the pull request.
+10. Changelog (Optional)
+It is sometimes difficult to keep track of changes made to an ontology. Some ontology teams opt to document changes in a changelog (simply a text file in your repository) so that when release day comes, you know everything you have changed. This is advisable at least for major changes (such as a new release system, a new pattern or template etc.).
+ + + + + + +Updating the Documentation
+The documentation for COB is managed in two places (relative to the repository root):
+-
+
- The
docsdirectory contains all the files that pertain to the content of the documentation (more below)
+ - the
mkdocs.yamlfile contains the documentation config, in particular its navigation bar and theme.
+
The documentation is hosted using GitHub pages, on a special branch of the repository (called gh-pages). It is important that this branch is never deleted - it contains all the files GitHub pages needs to render and deploy the site. It is also important to note that the gh-pages branch should never be edited manually. All changes to the docs happen inside the docs directory on the main branch.
Editing the docs
+Changing content
+All the documentation is contained in the docs directory, and is managed in Markdown. Markdown is a very simple and convenient way to produce text documents with formatting instructions, and is very easy to learn - it is also used, for example, in GitHub issues. This is a normal editing workflow:
-
+
- Open the
.mdfile you want to change in an editor of choice (a simple text editor is often best). IMPORTANT: Do not edit any files in thedocs/odk-workflows/directory. These files are managed by the ODK system and will be overwritten when the repository is upgraded! If you wish to change these files, make an issue on the ODK issue tracker.
+ - Perform the edit and save the file +
- Commit the file to a branch, and create a pull request as usual. +
- If your development team likes your changes, merge the docs into master branch. +
- Deploy the documentation (see below) +
Deploy the documentation
+The documentation is not automatically updated from the Markdown, and needs to be deployed deliberately. To do this, perform the following steps:
+-
+
- In your terminal, navigate to the edit directory of your ontology, e.g.:
+
cd cob/src/ontology
+ - Now you are ready to build the docs as follows:
+
sh run.sh make update_docs+ Mkdocs now sets off to build the site from the markdown pages. You will be asked to-
+
- Enter your username +
- Enter your password (see here for using GitHub access tokens instead) + IMPORTANT: Using password based authentication will be deprecated this year (2021). Make sure you read up on personal access tokens if that happens! +
+
If everything was successful, you will see a message similar to this one:
+INFO - Your documentation should shortly be available at: https://OBOFoundry.github.io/COB/
+3. Just to double check, you can now navigate to your documentation pages (usually https://OBOFoundry.github.io/COB/).
+ Just make sure you give GitHub 2-5 minutes to build the pages!
The release workflow
+The release workflow recommended by the ODK is based on GitHub releases and works as follows:
+-
+
- Run a release with the ODK +
- Review the release +
- Merge to main branch +
- Create a GitHub release +
These steps are outlined in detail in the following.
+Run a release with the ODK
+Preparation:
+-
+
- Ensure that all your pull requests are merged into your main (master) branch +
- Make sure that all changes to master are committed to GitHub (
git statusshould say that there are no modified files)
+ - Locally make sure you have the latest changes from master (
git pull)
+ - Checkout a new branch (e.g.
git checkout -b release-2021-01-01)
+ - You may or may not want to refresh your imports as part of your release strategy (see here) +
- Make sure you have the latest ODK installed by running
docker pull obolibrary/odkfull
+
To actually run the release, you:
+-
+
- Open a command line terminal window and navigate to the src/ontology directory (
cd cob/src/ontology)
+ - Run release pipeline:
sh run.sh make prepare_release -B. Note that for some ontologies, this process can take up to 90 minutes - especially if there are large ontologies you depend on, like PRO or CHEBI.
+ - If everything went well, you should see the following output on your machine:
Release files are now in ../.. - now you should commit, push and make a release on your git hosting site such as GitHub or GitLab.
+
This will create all the specified release targets (OBO, OWL, JSON, and the variants, ont-full and ont-base) and copy them into your release directory (the top level of your repo).
+Review the release
+-
+
- (Optional) Rough check. This step is frequently skipped, but for the more paranoid among us (like the author of this doc), this is a 3 minute additional effort for some peace of mind. Open the main release (cob.owl) in you favourite development environment (i.e. Protégé) and eyeball the hierarchy. We recommend two simple checks:
-
+
- Does the very top level of the hierarchy look ok? This means that all new terms have been imported/updated correctly. +
- Does at least one change that you know should be in this release appear? For example, a new class. This means that the release was actually based on the recent edit file. +
+ - Commit your changes to the branch and make a pull request +
- In your GitHub pull request, review the following three files in detail (based on our experience):
-
+
cob.obo- this reflects a useful subset of the whole ontology (everything that can be covered by OBO format). OBO format has that speaking for it: it is very easy to review!
+cob-base.owl- this reflects the asserted axioms in your ontology that you have actually edited.
+- Ideally also take a look at
cob-full.owl, which may reveal interesting new inferences you did not know about. Note that the diff of this file is sometimes quite large.
+
+ - Like with every pull request, we recommend to always employ a second set of eyes when reviewing a PR! +
Merge the main branch
+Once your CI checks have passed, and your reviews are completed, you can now merge the branch into your main branch (don't forget to delete the branch afterwards - a big button will appear after the merge is finished).
+Create a GitHub release
+-
+
- Go to your releases page on GitHub by navigating to your repository, and then clicking on releases (usually on the right, for example: https://github.com/OBOFoundry/COB/releases). Then click "Draft new release" +
- As the tag version you need to choose the date on which your ontologies were build. You can find this, for example, by looking at the
cob.obofile and check thedata-version:property. The date needs to be prefixed with av, so, for examplev2020-02-06.
+ - You can write whatever you want in the release title, but we typically write the date again. The description underneath should contain a concise list of changes or term additions. +
- Click "Publish release". Done. +
Debugging typical ontology release problems
+Problems with memory
+When you are dealing with large ontologies, you need a lot of memory. When you see error messages relating to large ontologies such as CHEBI, PRO, NCBITAXON, or Uberon, you should think of memory first, see here.
+Problems when using OBO format based tools
+Sometimes you will get cryptic error messages when using legacy tools using OBO format, such as the ontology release tool (OORT), which is also available as part of the ODK docker container. In these cases, you need to track down what axiom or annotation actually caused the breakdown. In our experience (in about 60% of the cases) the problem lies with duplicate annotations (def, comment) which are illegal in OBO. Here is an example recipe of how to deal with such a problem:
-
+
- If you get a message like
make: *** [cl.Makefile:84: oort] Error 255you might have a OORT error.
+ - To debug this, in your terminal enter
sh run.sh make IMP=false PAT=false oort -B(assuming you are already in the ontology folder in your directory)
+ - This should show you where the error is in the log (eg multiple different definitions) +WARNING: THE FIX BELOW IS NOT IDEAL, YOU SHOULD ALWAYS TRY TO FIX UPSTREAM IF POSSIBLE +
- Open
cob-edit.owlin Protégé and find the offending term and delete all offending issue (e.g. delete ALL definition, if the problem was "multiple def tags not allowed") and save. +*While this is not idea, as it will remove all definitions from that term, it will be added back again when the term is fixed in the ontology it was imported from and added back in.
+ - Rerun
sh run.sh make IMP=false PAT=false oort -Band if it all passes, commit your changes to a branch and make a pull request as usual.
+
Managing your ODK repository
+Updating your ODK repository
+Your ODK repositories configuration is managed in src/ontology/cob-odk.yaml. Once you have made your changes, you can run the following to apply your changes to the repository:
sh run.sh make update_repo
+There are a large number of options that can be set to configure your ODK, but we will only discuss a few of them here.
+NOTE for Windows users:
+You may get a cryptic failure such as Set Illegal Option - if the update script located in src/scripts/update_repo.sh
+was saved using Windows Line endings. These need to change to unix line endings. In Notepad++, for example, you can
+click on Edit->EOL Conversion->Unix LF to change this.
Managing imports
+You can use the update repository workflow described on this page to perform the following operations to your imports:
+-
+
- Add a new import +
- Modify an existing import +
- Remove an import you no longer want +
- Customise an import +
We will discuss all these workflows in the following.
+Add new import
+To add a new import, you first edit your odk config as described above, adding an id to the product list in the import_group section (for the sake of this example, we assume you already import RO, and your goal is to also import GO):
import_group:
+ products:
+ - id: ro
+ - id: go
+Note: our ODK file should only have one import_group which can contain multiple imports (in the products section). Next, you run the update repo workflow to apply these changes. Note that by default, this module is going to be a SLME Bottom module, see here. To change that or customise your module, see section "Customise an import". To finalise the addition of your import, perform the following steps:
-
+
- Add an import statement to your
src/ontology/cob-edit.owlfile. We suggest to do this using a text editor, by simply copying an existing import declaration and renaming it to the new ontology import, for example as follows: +... + Ontology(<http://purl.obolibrary.org/obo/cob.owl> + Import(<http://purl.obolibrary.org/obo/cob/imports/ro_import.owl>) + Import(<http://purl.obolibrary.org/obo/cob/imports/go_import.owl>) + ...
+ - Add your imports redirect to your catalog file
src/ontology/catalog-v001.xml, for example: +<uri name="http://purl.obolibrary.org/obo/cob/imports/go_import.owl" uri="imports/go_import.owl"/>
+ - Test whether everything is in order:
-
+
- Refresh your import +
- Open in your Ontology Editor of choice (Protege) and ensure that the expected terms are imported. +
+
Note: The catalog file src/ontology/catalog-v001.xml has one purpose: redirecting
+imports from URLs to local files. For example, if you have
Import(<http://purl.obolibrary.org/obo/cob/imports/go_import.owl>)
+in your editors file (the ontology) and
+<uri name="http://purl.obolibrary.org/obo/cob/imports/go_import.owl" uri="imports/go_import.owl"/>
+in your catalog, tools like robot or Protégé will recognize the statement
+in the catalog file to redirect the URL http://purl.obolibrary.org/obo/cob/imports/go_import.owl
+to the local file imports/go_import.owl (which is in your src/ontology directory).
Modify an existing import
+If you simply wish to refresh your import in light of new terms, see here. If you wish to change the type of your module see section "Customise an import".
+Remove an existing import
+To remove an existing import, perform the following steps:
+-
+
- remove the import declaration from your
src/ontology/cob-edit.owl.
+ - remove the id from your
src/ontology/cob-odk.yaml, eg.- id: gofrom the list ofproductsin theimport_group.
+ - run update repo workflow +
- delete the associated files manually:
-
+
src/imports/go_import.owl
+src/imports/go_terms.txt
+
+ - Remove the respective entry from the
src/ontology/catalog-v001.xmlfile.
+
Customise an import
+By default, an import module extracted from a source ontology will be a SLME module, see here. There are various options to change the default.
+The following change to your repo config (src/ontology/cob-odk.yaml) will switch the go import from an SLME module to a simple ROBOT filter module:
import_group:
+ products:
+ - id: ro
+ - id: go
+ module_type: filter
+A ROBOT filter module is, essentially, importing all external terms declared by your ontology (see here on how to declare external terms to be imported). Note that the filter module does
+not consider terms/annotations from namespaces other than the base-namespace of the ontology itself. For example, in the
+example of GO above, only annotations / axioms related to the GO base IRI (http://purl.obolibrary.org/obo/GO_) would be considered. This
+behaviour can be changed by adding additional base IRIs as follows:
import_group:
+ products:
+ - id: go
+ module_type: filter
+ base_iris:
+ - http://purl.obolibrary.org/obo/GO_
+ - http://purl.obolibrary.org/obo/CL_
+ - http://purl.obolibrary.org/obo/BFO
+If you wish to customise your import entirely, you can specify your own ROBOT command to do so. To do that, add the following to your repo config (src/ontology/cob-odk.yaml):
import_group:
+ products:
+ - id: ro
+ - id: go
+ module_type: custom
+Now add a new goal in your custom Makefile (src/ontology/cob.Makefile, not src/ontology/Makefile).
imports/go_import.owl: mirror/ro.owl imports/ro_terms_combined.txt
+ if [ $(IMP) = true ]; then $(ROBOT) query -i $< --update ../sparql/preprocess-module.ru \
+ extract -T imports/ro_terms_combined.txt --force true --individuals exclude --method BOT \
+ query --update ../sparql/inject-subset-declaration.ru --update ../sparql/postprocess-module.ru \
+ annotate --ontology-iri $(ONTBASE)/$@ $(ANNOTATE_ONTOLOGY_VERSION) --output $@.tmp.owl && mv $@.tmp.owl $@; fi
+Now feel free to change this goal to do whatever you wish it to do! It probably makes some sense (albeit not being a strict necessity), to leave most of the goal instead and replace only:
+extract -T imports/ro_terms_combined.txt --force true --individuals exclude --method BOT \
+to another ROBOT pipeline.
+Add a component
+A component is an import which belongs to your ontology, e.g. is managed by +you and your team.
+-
+
- Open
src/ontology/cob-odk.yaml
+ - If you dont have it yet, add a new top level section
components
+ - Under the
componentssection, add a new section calledproducts. +This is where all your components are specified
+ - Under the
productssection, add a new component, e.g.- filename: mycomp.owl
+
Example
+components:
+ products:
+ - filename: mycomp.owl
+When running sh run.sh make update_repo, a new file src/ontology/components/mycomp.owl will
+be created which you can edit as you see fit. Typical ways to edit:
-
+
- Using a ROBOT template to generate the component (see below) +
- Manually curating the component separately with Protégé or any other editor +
- Providing a
components/mycomp.owl:make target insrc/ontology/cob.Makefile+and provide a custom command to generate the component-
+
WARNING: Note that the custom rule to generate the component MUST NOT depend on any other ODK-generated file such as seed files and the like (see issue).
+
+ - Providing an additional attribute for the component in
src/ontology/cob-odk.yaml,source, +to specify that this component should simply be downloaded from somewhere on the web.
+
Adding a new component based on a ROBOT template
+Since ODK 1.3.2, it is possible to simply link a ROBOT template to a component without having to specify any of the import logic. In order to add a new component that is connected to one or more template files, follow these steps:
+-
+
- Open
src/ontology/cob-odk.yaml.
+ - Make sure that
use_templates: TRUEis set in the global project options. You should also make sure thatuse_context: TRUEis set in case you are using prefixes in your templates that are not known torobot, such asOMOP:,CPONT:and more. All non-standard prefixes you are using should be added toconfig/context.json.
+ - Add another component to the
productssection.
+ - To activate this component to be template-driven, simply say:
use_template: TRUE. This will create an empty template for you in the templates directory, which will automatically be processed when recreating the component (e.g.run.bat make recreate-mycomp).
+ - If you want to use more than one component, use the
templatesfield to add as many template names as you wish. ODK will look for them in thesrc/templatesdirectory.
+ - Advanced: If you want to provide additional processing options, you can use the
template_optionsfield. This should be a string with option from robot template. One typical example for additional options you may want to provide is--add-prefixes config/context.jsonto ensure the prefix map of your context is provided torobot, see above.
+
Example:
+components:
+ products:
+ - filename: mycomp.owl
+ use_template: TRUE
+ template_options: --add-prefixes config/context.json
+ templates:
+ - template1.tsv
+ - template2.tsv
+Note: if your mirror is particularly large and complex, read this ODK recommendation.
+ + + + + + +Repository structure
+The main kinds of files in the repository:
+-
+
- Release files +
- Imports +
- Components +
Release files
+Release file are the file that are considered part of the official ontology release and to be used by the community. A detailed description of the release artefacts can be found here.
+Imports
+Imports are subsets of external ontologies that contain terms and axioms you would like to re-use in your ontology. These are considered "external", like dependencies in software development, and are not included in your "base" product, which is the release artefact which contains only those axioms that you personally maintain.
+These are the current imports in COB
+| Import | +URL | +Type | +
|---|---|---|
| ro | +http://purl.obolibrary.org/obo/ro.owl | +slme | +
| omo | +http://purl.obolibrary.org/obo/omo.owl | +mirror | +
Components
+Components, in contrast to imports, are considered full members of the ontology. This means that any axiom in a component is also included in the ontology base - which means it is considered native to the ontology. While this sounds complicated, consider this: conceptually, no component should be part of more than one ontology. If that seems to be the case, we are most likely talking about an import. Components are often not needed for ontologies, but there are some use cases:
+-
+
- There is an automated process that generates and re-generates a part of the ontology +
- A part of the ontology is managed in ROBOT templates +
- The expressivity of the component is higher than the format of the edit file. For example, people still choose to manage their ontology in OBO format (they should not) missing out on a lot of owl features. They may choose to manage logic that is beyond OBO in a specific OWL component. +
These are the components in COB
+| Filename | +URL | +
|---|---|
| cob-annotations.owl | +None | +
| cob-to-external.owl | +None | +
Setting up your Docker environment for ODK use
+One of the most frequent problems with running the ODK for the first time is failure because of lack of memory. This can look like a Java OutOfMemory exception,
+but more often than not it will appear as something like an Error 137. There are two places you need to consider to set your memory:
-
+
- Your src/ontology/run.sh (or run.bat) file. You can set the memory in there by adding
+
robot_java_args: '-Xmx8G'to your src/ontology/cob-odk.yaml file, see for example here.
+ - Set your docker memory. By default, it should be about 10-20% more than your
robot_java_argsvariable. You can manage your memory settings +by right-clicking on the docker whale in your system bar-->Preferences-->Resources-->Advanced, see picture below.
+

Update Imports Workflow
+This page discusses how to update the contents of your imports, like adding or removing terms. If you are looking to customise imports, like changing the module type, see here.
+Importing a new term
+Note: some ontologies now use a merged-import system to manage dynamic imports, for these please follow instructions in the section title "Using the Base Module approach".
+Importing a new term is split into two sub-phases:
+-
+
- Declaring the terms to be imported +
- Refreshing imports dynamically +
Declaring terms to be imported
+There are three ways to declare terms that are to be imported from an external ontology. Choose the appropriate one for your particular scenario (all three can be used in parallel if need be):
+-
+
- Protégé-based declaration +
- Using term files +
- Using the custom import template +
Protégé-based declaration
+This workflow is to be avoided, but may be appropriate if the editor does not have access to the ODK docker container. +This approach also applies to ontologies that use base module import approach.
+-
+
- Open your ontology (edit file) in Protégé (5.5+). +
- Select 'owl:Thing' +
- Add a new class as usual. +
- Paste the full iri in the 'Name:' field, for example, http://purl.obolibrary.org/obo/CHEBI_50906. +
- Click 'OK' +
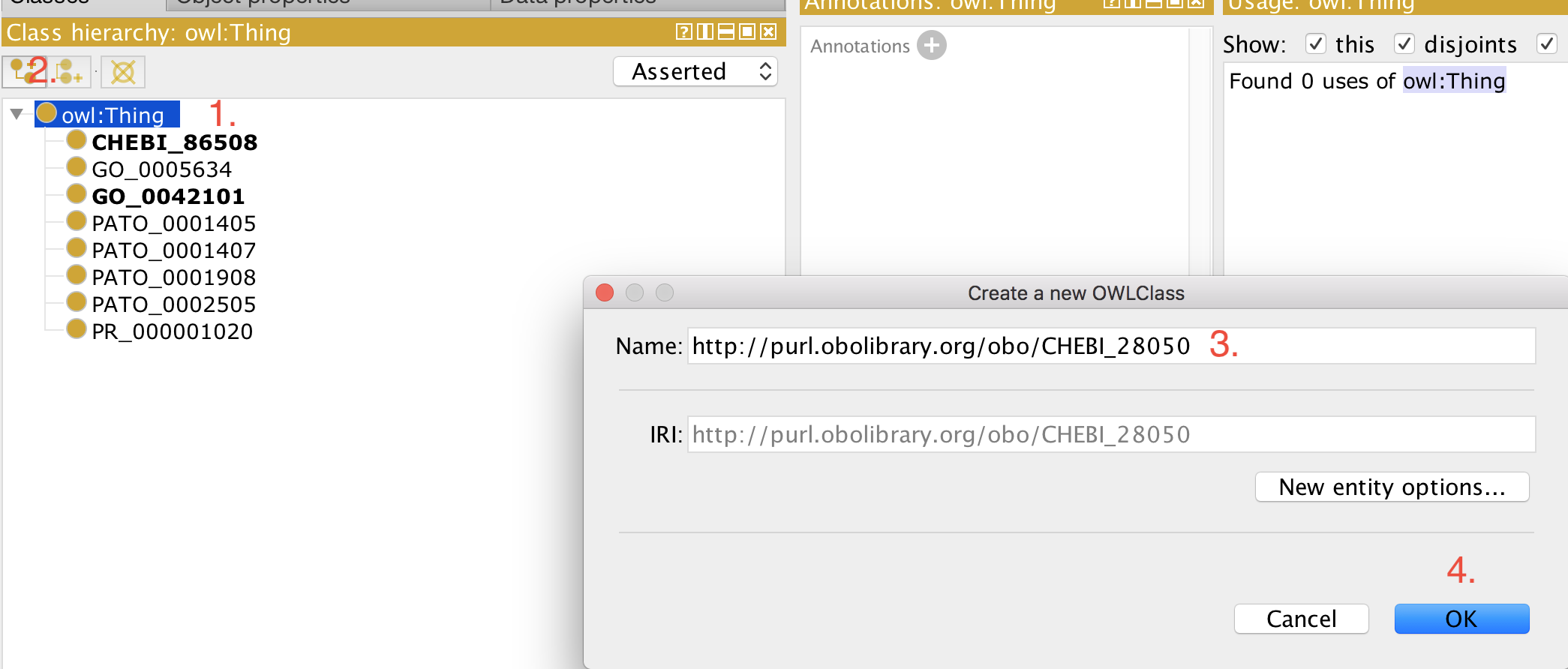
Now you can use this term for example to construct logical definitions. The next time the imports are refreshed (see how to refresh here), the metadata (labels, definitions, etc.) for this term are imported from the respective external source ontology and becomes visible in your ontology.
+Using term files
+Every import has, by default a term file associated with it, which can be found in the imports directory. For example, if you have a GO import in src/ontology/go_import.owl, you will also have an associated term file src/ontology/go_terms.txt. You can add terms in there simply as a list:
GO:0008150
+GO:0008151
+Now you can run the refresh imports workflow) and the two terms will be imported.
+Using the custom import template
+This workflow is appropriate if:
+-
+
- You prefer to manage all your imported terms in a single file (rather than multiple files like in the "Using term files" workflow above). +
- You wish to augment your imported ontologies with additional information. This requires a cautionary discussion. +
To enable this workflow, you add the following to your ODK config file (src/ontology/cob-odk.yaml), and update the repository:
use_custom_import_module: TRUE
+Now you can manage your imported terms directly in the custom external terms template, which is located at src/templates/external_import.owl. Note that this file is a ROBOT template, and can, in principle, be extended to include any axioms you like. Before extending the template, however, read the following carefully.
The main purpose of the custom import template is to enable the management off all terms to be imported in a centralised place. To enable that, you do not have to do anything other than maintaining the template. So if you, say currently import APOLLO_SV:00000480, and you wish to import APOLLO_SV:00000532, you simply add a row like this:
ID Entity Type
+ID TYPE
+APOLLO_SV:00000480 owl:Class
+APOLLO_SV:00000532 owl:Class
+When the imports are refreshed see imports refresh workflow, the term(s) will simply be imported from the configured ontologies.
+Now, if you wish to extend the Makefile (which is beyond these instructions) and add, say, synonyms to the imported terms, you can do that, but you need to (a) preserve the ID and ENTITY columns and (b) ensure that the ROBOT template is valid otherwise, see here.
WARNING. Note that doing this is a widespread antipattern (see related issue). You should not change the axioms of terms that do not belong into your ontology unless necessary - such changes should always be pushed into the ontology where they belong. However, since people are doing it, whether the OBO Foundry likes it or not, at least using the custom imports module as described here localises the changes to a single simple template and ensures that none of the annotations added this way are merged into the base file.
+Refresh imports
+If you want to refresh the import yourself (this may be necessary to pass the travis tests), and you have the ODK installed, you can do the following (using go as an example):
+First, you navigate in your terminal to the ontology directory (underneath src in your hpo root directory).
+cd src/ontology
+Then, you regenerate the import that will now include any new terms you have added. Note: You must have docker installed.
+sh run.sh make PAT=false imports/go_import.owl -B
+Since ODK 1.2.27, it is also possible to simply run the following, which is the same as the above:
+sh run.sh make refresh-go
+Note that in case you changed the defaults, you need to add IMP=true and/or MIR=true to the command below:
sh run.sh make IMP=true MIR=true PAT=false imports/go_import.owl -B
+If you wish to skip refreshing the mirror, i.e. skip downloading the latest version of the source ontology for your import (e.g. go.owl for your go import) you can set MIR=false instead, which will do the exact same thing as the above, but is easier to remember:
sh run.sh make IMP=true MIR=false PAT=false imports/go_import.owl -B
+Using the Base Module approach
+Since ODK 1.2.31, we support an entirely new approach to generate modules: Using base files. +The idea is to only import axioms from ontologies that actually belong to it. +A base file is a subset of the ontology that only contains those axioms that nominally +belong there. In other words, the base file does not contain any axioms that belong +to another ontology. An example would be this:
+Imagine this being the full Uberon ontology:
+Axiom 1: BFO:123 SubClassOf BFO:124
+Axiom 1: UBERON:123 SubClassOf BFO:123
+Axiom 1: UBERON:124 SubClassOf UBERON 123
+The base file is the set of all axioms that are about UBERON terms:
+Axiom 1: UBERON:123 SubClassOf BFO:123
+Axiom 1: UBERON:124 SubClassOf UBERON 123
+I.e.
+Axiom 1: BFO:123 SubClassOf BFO:124
+Gets removed.
+The base file pipeline is a bit more complex than the normal pipelines, because +of the logical interactions between the imported ontologies. This is solved by _first +merging all mirrors into one huge file and then extracting one mega module from it.
+Example: Let's say we are importing terms from Uberon, GO and RO in our ontologies. +When we use the base pipelines, we
+1) First obtain the base (usually by simply downloading it, but there is also an option now to create it with ROBOT)
+2) We merge all base files into one big pile
+3) Then we extract a single module imports/merged_import.owl
The first implementation of this pipeline is PATO, see https://github.com/pato-ontology/pato/blob/master/src/ontology/pato-odk.yaml.
+To check if your ontology uses this method, check src/ontology/cob-odk.yaml to see if use_base_merging: TRUE is declared under import_group
If your ontology uses Base Module approach, please use the following steps:
+First, add the term to be imported to the term file associated with it (see above "Using term files" section if this is not clear to you)
+Next, you navigate in your terminal to the ontology directory (underneath src in your hpo root directory).
+cd src/ontology
+Then refresh imports by running
+sh run.sh make imports/merged_import.owl
+Note: if your mirrors are updated, you can run sh run.sh make no-mirror-refresh-merged
This requires quite a bit of memory on your local machine, so if you encounter an error, it might be a lack of memory on your computer. A solution would be to create a ticket in an issue tracker requesting for the term to be imported, and one of the local devs should pick this up and run the import for you.
+Lastly, restart Protégé, and the term should be imported in ready to be used.
+ + + + + + +Adding components to an ODK repo
+For details on what components are, please see component section of repository file structure document.
+To add custom components to an ODK repo, please follow the following steps:
+1) Locate your odk yaml file and open it with your favourite text editor (src/ontology/cob-odk.yaml) +2) Search if there is already a component section to the yaml file, if not add it accordingly, adding the name of your component:
+components:
+ products:
+ - filename: your-component-name.owl
+3) Add the component to your catalog file (src/ontology/catalog-v001.xml)
+ <uri name="http://purl.obolibrary.org/obo/cob/components/your-component-name.owl" uri="components/your-component-name.owl"/>
+4) Add the component to the edit file (src/ontology/cob-edit.obo) +for .obo formats:
+import: http://purl.obolibrary.org/obo/cob/components/your-component-name.owl
+for .owl formats:
+Import(<http://purl.obolibrary.org/obo/cob/components/your-component-name.owl>)
+5) Refresh your repo by running sh run.sh make update_repo - this should create a new file in src/ontology/components.
+6) In your custom makefile (src/ontology/cob.Makefile) add a goal for your custom make file. In this example, the goal is a ROBOT template.
$(COMPONENTSDIR)/your-component-name.owl: $(SRC) ../templates/your-component-template.tsv
+ $(ROBOT) template --template ../templates/your-component-template.tsv \
+ annotate --ontology-iri $(ONTBASE)/$@ --output $(COMPONENTSDIR)/your-component-name.owl
+(If using a ROBOT template, do not forget to add your template tsv in src/templates/)
+7) Make the file by running sh run.sh make components/your-component-name.owl
Default ODK Workflows
+ + + + + + + +'+ escapeHtml(title) + '
' + escapeHtml(summary) +'
' + noResultsText + '
'); + } +} + +function doSearch () { + var query = document.getElementById('mkdocs-search-query').value; + if (query.length > min_search_length) { + if (!window.Worker) { + displayResults(search(query)); + } else { + searchWorker.postMessage({query: query}); + } + } else { + // Clear results for short queries + displayResults([]); + } +} + +function initSearch () { + var search_input = document.getElementById('mkdocs-search-query'); + if (search_input) { + search_input.addEventListener("keyup", doSearch); + } + var term = getSearchTermFromLocation(); + if (term) { + search_input.value = term; + doSearch(); + } +} + +function onWorkerMessage (e) { + if (e.data.allowSearch) { + initSearch(); + } else if (e.data.results) { + var results = e.data.results; + displayResults(results); + } else if (e.data.config) { + min_search_length = e.data.config.min_search_length-1; + } +} + +if (!window.Worker) { + console.log('Web Worker API not supported'); + // load index in main thread + $.getScript(joinUrl(base_url, "search/worker.js")).done(function () { + console.log('Loaded worker'); + init(); + window.postMessage = function (msg) { + onWorkerMessage({data: msg}); + }; + }).fail(function (jqxhr, settings, exception) { + console.error('Could not load worker.js'); + }); +} else { + // Wrap search in a web worker + var searchWorker = new Worker(joinUrl(base_url, "search/worker.js")); + searchWorker.postMessage({init: true}); + searchWorker.onmessage = onWorkerMessage; +} diff --git a/search/search_index.json b/search/search_index.json new file mode 100644 index 0000000..ed4a7c4 --- /dev/null +++ b/search/search_index.json @@ -0,0 +1 @@ +{"config":{"indexing":"full","lang":["en"],"min_search_length":3,"prebuild_index":false,"separator":"[\\s\\-]+"},"docs":[{"location":"","text":"COB Ontology Documentation This project is an attempt to bring together key terms from a wide range of Open Biological and Biomedical Ontology (OBO) projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms. Our plan is to keep this ontology small, but ensure that one or more COB terms can be used as the root of any given OBO library ontology. Editors Guide You can find descriptions of the standard ontology engineering workflows here . Issue Tracker If you are responsible for an OBO ontology you can see any tickets that pertain to your ontology by looking for the label with your ontology ID. For example: GO : https://github.com/OBOFoundry/COB/labels/GO OBI : https://github.com/OBOFoundry/COB/labels/OBI","title":"Getting started"},{"location":"#cob-ontology-documentation","text":"This project is an attempt to bring together key terms from a wide range of Open Biological and Biomedical Ontology (OBO) projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms. Our plan is to keep this ontology small, but ensure that one or more COB terms can be used as the root of any given OBO library ontology.","title":"COB Ontology Documentation"},{"location":"#editors-guide","text":"You can find descriptions of the standard ontology engineering workflows here .","title":"Editors Guide"},{"location":"#issue-tracker","text":"If you are responsible for an OBO ontology you can see any tickets that pertain to your ontology by looking for the label with your ontology ID. For example: GO : https://github.com/OBOFoundry/COB/labels/GO OBI : https://github.com/OBOFoundry/COB/labels/OBI","title":"Issue Tracker"},{"location":"cite/","text":"How to cite COB","title":"Cite"},{"location":"cite/#how-to-cite-cob","text":"","title":"How to cite COB"},{"location":"contributing/","text":"How to contribute to COB","title":"Contributing"},{"location":"contributing/#how-to-contribute-to-cob","text":"","title":"How to contribute to COB"},{"location":"editors/","text":"Editors documentation Do not edit cob.owl directly. Instead edit cob-edit.owl Do not edit cob-to-external.owl directly. Instead edit the tsv and run the Makefile. Run a COB release: See standard ODK release docs here . Run COB integration tests: All COB tests: cd src/ontology sh run.sh make cob_test Basic COB tests: cd src/ontology sh run.sh make main_test Run regular ontology QC tests: cd src/ontology sh run.sh make IMP=false test","title":"Editors Readme"},{"location":"editors/#editors-documentation","text":"Do not edit cob.owl directly. Instead edit cob-edit.owl Do not edit cob-to-external.owl directly. Instead edit the tsv and run the Makefile.","title":"Editors documentation"},{"location":"editors/#run-a-cob-release","text":"See standard ODK release docs here .","title":"Run a COB release:"},{"location":"editors/#run-cob-integration-tests","text":"","title":"Run COB integration tests:"},{"location":"editors/#all-cob-tests","text":"cd src/ontology sh run.sh make cob_test","title":"All COB tests:"},{"location":"editors/#basic-cob-tests","text":"cd src/ontology sh run.sh make main_test","title":"Basic COB tests:"},{"location":"editors/#run-regular-ontology-qc-tests","text":"cd src/ontology sh run.sh make IMP=false test","title":"Run regular ontology QC tests:"},{"location":"exploring/","text":"Exploring COB Browse COB is available on the following browsers: Ontobee OLS BioPortal You can also enter any COB class PURL into a browser to resolve it; e.g. http://purl.obolibrary.org/obo/COB_0000011 (atom) Files End products cob.owl : the current version of COB containing new terms in the COB namespace cob-to-external.owl COB terms and OBO Foundry counterparts with proposed equivalencies cob-examples.owl : additional child terms as examples. This file is edited by hand. Editors/Source files cob-edit.owl : the editors version cob-to-external.tsv : TSV source of cob-to-external Makefile : workflow for build COB COB demo As a demonstration of how COB could be used to unify OBO ontologies in the future we produce an ontology: products/demo-cob.owl This has selected subsets of certain ontologies merged in with cob-to-external. It is incomplete and messy. Please see the Makefile for how to add more to it.","title":"Exploring COB"},{"location":"exploring/#exploring-cob","text":"","title":"Exploring COB"},{"location":"exploring/#browse","text":"COB is available on the following browsers: Ontobee OLS BioPortal You can also enter any COB class PURL into a browser to resolve it; e.g. http://purl.obolibrary.org/obo/COB_0000011 (atom)","title":"Browse"},{"location":"exploring/#files","text":"End products cob.owl : the current version of COB containing new terms in the COB namespace cob-to-external.owl COB terms and OBO Foundry counterparts with proposed equivalencies cob-examples.owl : additional child terms as examples. This file is edited by hand. Editors/Source files cob-edit.owl : the editors version cob-to-external.tsv : TSV source of cob-to-external Makefile : workflow for build COB","title":"Files"},{"location":"exploring/#cob-demo","text":"As a demonstration of how COB could be used to unify OBO ontologies in the future we produce an ontology: products/demo-cob.owl This has selected subsets of certain ontologies merged in with cob-to-external. It is incomplete and messy. Please see the Makefile for how to add more to it.","title":"COB demo"},{"location":"history/","text":"A brief history of COB The following page gives an overview of the history of COB.","title":"History"},{"location":"history/#a-brief-history-of-cob","text":"The following page gives an overview of the history of COB.","title":"A brief history of COB"},{"location":"integration-tests/","text":"Integration Tests Currently we have two kinds: Coherency : testing if an ontology is coherent when combined with COB + ext Coverage : testing if an ontology has classes that are not subclasses (direct or indirect) of COB classes See the products folder","title":"Integration Tests"},{"location":"integration-tests/#integration-tests","text":"Currently we have two kinds: Coherency : testing if an ontology is coherent when combined with COB + ext Coverage : testing if an ontology has classes that are not subclasses (direct or indirect) of COB classes See the products folder","title":"Integration Tests"},{"location":"obo-bridge/","text":"Placing OBO ontologies under COB COB is intended as a central place where different ontologies within OBO can discuss and agree where the different root terms across OBO belong in the COB hierarchy. The source of truth for this is a file \"COB to External\" COB to External The source for the file is in the src/ontology/components folder in GitHub: cob-to-external.tsv The file is in SSSOM tsv format. Example entries The following entries use the owl:equivalentClass predicate to indicate that the COB IDs are equivalent to the the corresponding OBO concept. subject_id subject_label predicate_id object_id object_label notes COB:0000003 mass owl:equivalentClass PATO:0000125 mass . COB:0000004 charge owl:equivalentClass PATO:0002193 electric . COB:0000006 material entity owl:equivalentClass BFO:0000040 material entity . COB:0000008 proton owl:equivalentClass CHEBI:24636 proton . COB:0000017 cell owl:equivalentClass CL:0000000 cell . COB:0000026 processed material entity owl:equivalentClass OBI:0000047 processed material . COB:0000031 immaterial entity owl:equivalentClass BFO:0000041 immaterial entity . COB:0000033 realizable owl:equivalentClass BFO:0000017 realizable entity . Multiple OBO IDs for one COB ID Ideally no COB ID is equivalent to more than one OBO ID, but in practice different ontologies have minted IDs for what turned out to be the same concept. COB serves as a useful clearinghouse for making these lack of orthogonality transparent. For example, the COB concept of \"organism\" (which includes viruses): subject_id subject_label predicate_id object_id object_label notes COB:0000022 organism owl:equivalentClass NCBITaxon:1 root . COB:0000022 organism owl:equivalentClass OBI:0100026 organism . COB:0000022 organism owl:equivalentClass CARO:0001010 organism or virus or viroid . subClass links we can also declare the OBO class to be a subclass of the COB class. For example, the concept of a cell in the plant ontology is the species equivalent of the species-generic COB cell, so formally it is a subclass: subject_id subject_label predicate_id object_id object_label notes COB:0000017 cell owl:equivalentClass CL:0000000 cell . COB:0000017 cell sssom:superClassOf PO:0009002 plant cell . COB:0000018 native cell owl:equivalentClass CL:0000003 native cell . COB:0000018 native cell sssom:superClassOf XAO:0003012 xenopus cell . COB:0000018 native cell sssom:superClassOf ZFA:0009000 zebrafish cell . COB:0000018 native cell sssom:superClassOf PO:0025606 native plant cell . COB:0000019 cell in vitro owl:equivalentClass CL:0001034 cell in vitro . Similarly, we can record the fact that we are committing to group SO entities as material entities subject_id subject_label predicate_id object_id object_label notes COB:0000006 material entity sssom:superClassOf SO:0000110 sequence_feature . COB:0000006 material entity sssom:superClassOf SO:0001060 sequence_variant . COB:0000006 material entity sssom:superClassOf SO:0001260 sequence_collection . Formal lack of commitment We can record a formal lack of commitment in cob-to-external: subject_id subject_label predicate_id object_id object_label notes COB:0000011 atom skos:closeMatch CHEBI:33250 atom . COB:0000013 molecular entity skos:closeMatch CHEBI:23367 molecular entity No exact match in CHEBI COB:0000013 molecular entity skos:closeMatch CHEBI:25367 molecule This is electrically neutral in CHEBI but in COB it is generic. We should make it union of molecule and polyatomic ion We cannot make these equivalent because we would end up with incoherencies in COB Proposing changes to cob-to-external Anyone is welcome to make Pull Requests on the cob-to-external file. The pull request will be discussed openly and transparently, and anyone can make comments on the GitHub PR","title":"Placing OBO ontologies under COB"},{"location":"obo-bridge/#placing-obo-ontologies-under-cob","text":"COB is intended as a central place where different ontologies within OBO can discuss and agree where the different root terms across OBO belong in the COB hierarchy. The source of truth for this is a file \"COB to External\"","title":"Placing OBO ontologies under COB"},{"location":"obo-bridge/#cob-to-external","text":"The source for the file is in the src/ontology/components folder in GitHub: cob-to-external.tsv The file is in SSSOM tsv format.","title":"COB to External"},{"location":"obo-bridge/#example-entries","text":"The following entries use the owl:equivalentClass predicate to indicate that the COB IDs are equivalent to the the corresponding OBO concept. subject_id subject_label predicate_id object_id object_label notes COB:0000003 mass owl:equivalentClass PATO:0000125 mass . COB:0000004 charge owl:equivalentClass PATO:0002193 electric . COB:0000006 material entity owl:equivalentClass BFO:0000040 material entity . COB:0000008 proton owl:equivalentClass CHEBI:24636 proton . COB:0000017 cell owl:equivalentClass CL:0000000 cell . COB:0000026 processed material entity owl:equivalentClass OBI:0000047 processed material . COB:0000031 immaterial entity owl:equivalentClass BFO:0000041 immaterial entity . COB:0000033 realizable owl:equivalentClass BFO:0000017 realizable entity .","title":"Example entries"},{"location":"obo-bridge/#multiple-obo-ids-for-one-cob-id","text":"Ideally no COB ID is equivalent to more than one OBO ID, but in practice different ontologies have minted IDs for what turned out to be the same concept. COB serves as a useful clearinghouse for making these lack of orthogonality transparent. For example, the COB concept of \"organism\" (which includes viruses): subject_id subject_label predicate_id object_id object_label notes COB:0000022 organism owl:equivalentClass NCBITaxon:1 root . COB:0000022 organism owl:equivalentClass OBI:0100026 organism . COB:0000022 organism owl:equivalentClass CARO:0001010 organism or virus or viroid .","title":"Multiple OBO IDs for one COB ID"},{"location":"obo-bridge/#subclass-links","text":"we can also declare the OBO class to be a subclass of the COB class. For example, the concept of a cell in the plant ontology is the species equivalent of the species-generic COB cell, so formally it is a subclass: subject_id subject_label predicate_id object_id object_label notes COB:0000017 cell owl:equivalentClass CL:0000000 cell . COB:0000017 cell sssom:superClassOf PO:0009002 plant cell . COB:0000018 native cell owl:equivalentClass CL:0000003 native cell . COB:0000018 native cell sssom:superClassOf XAO:0003012 xenopus cell . COB:0000018 native cell sssom:superClassOf ZFA:0009000 zebrafish cell . COB:0000018 native cell sssom:superClassOf PO:0025606 native plant cell . COB:0000019 cell in vitro owl:equivalentClass CL:0001034 cell in vitro . Similarly, we can record the fact that we are committing to group SO entities as material entities subject_id subject_label predicate_id object_id object_label notes COB:0000006 material entity sssom:superClassOf SO:0000110 sequence_feature . COB:0000006 material entity sssom:superClassOf SO:0001060 sequence_variant . COB:0000006 material entity sssom:superClassOf SO:0001260 sequence_collection .","title":"subClass links"},{"location":"obo-bridge/#formal-lack-of-commitment","text":"We can record a formal lack of commitment in cob-to-external: subject_id subject_label predicate_id object_id object_label notes COB:0000011 atom skos:closeMatch CHEBI:33250 atom . COB:0000013 molecular entity skos:closeMatch CHEBI:23367 molecular entity No exact match in CHEBI COB:0000013 molecular entity skos:closeMatch CHEBI:25367 molecule This is electrically neutral in CHEBI but in COB it is generic. We should make it union of molecule and polyatomic ion We cannot make these equivalent because we would end up with incoherencies in COB","title":"Formal lack of commitment"},{"location":"obo-bridge/#proposing-changes-to-cob-to-external","text":"Anyone is welcome to make Pull Requests on the cob-to-external file. The pull request will be discussed openly and transparently, and anyone can make comments on the GitHub PR","title":"Proposing changes to cob-to-external"},{"location":"using-cob/","text":"Using COB: An ontology developers guide Intended audience: technical leads for individual OBO ontology projects This is a draft guide. There are many technical issues that need to be worked out to transition to COB. We strongly recommend you do work here on a branch, and if you have problems, make an issue in the COB tracker and link to your PR 1. Add your roots to the OBO bridge See Bridging COB to OBO for instructions on how to make sure your ontology's root terms are mapped to COB. If you are not bridged to COB, or don't understand the bridge file, then you are not ready to use it in the production releases of your ontology. 2. Use a COB import Here we assume you are familiar with standard OBO tools such as ROBOT and the ROBOT extract command, or you are using the ODK to make releases. We assume here that all of your imports are to modules extracted from Base files. If you are not doing this, or don't know what a Base file is, then we recommend waiting until the documentation around these is more mature before you attempt to switch to COB. If you are already using a BFO import, then you should modify your build process to instead use COB. If you are configuring an ontology for the first time using the ODK, then all you need to is declare cob as one of your input ontologies. You should do this in place of bfo-classes (don't worry, you will still get most back via COB). Troubleshooting Dangling BFO class IDs The most likely problem will be dangling BFO class IDs. These may come from two sources: subClassOf axioms in a base module from root nodes domain, range, or similar constraining axioms in RO The first case should be rare, because most ontologies are rooted in a COB class with a BFO equivalent. However, in some cases, ontologies will be rooted in either: a class in BFO that is beneath the shoreline in COB, e.g. fiat object part is used as a root in ENVO (for now; see https://github.com/EnvironmentOntology/envo/pull/1252) a class in BFO that is above the shoreline in COB, e.g. a domain ontology class that directly subclasses a high level BFO class like continuant. These should be rare to non-existent The second category is harder. See https://github.com/OBOFoundry/COB/issues/213 Using COB for end-users If you are not an ontology developer or directly involved in the construction of one more ontologies, then there is not much opportunity to \"use\" COB.","title":"Using COB"},{"location":"using-cob/#using-cob-an-ontology-developers-guide","text":"Intended audience: technical leads for individual OBO ontology projects This is a draft guide. There are many technical issues that need to be worked out to transition to COB. We strongly recommend you do work here on a branch, and if you have problems, make an issue in the COB tracker and link to your PR","title":"Using COB: An ontology developers guide"},{"location":"using-cob/#1-add-your-roots-to-the-obo-bridge","text":"See Bridging COB to OBO for instructions on how to make sure your ontology's root terms are mapped to COB. If you are not bridged to COB, or don't understand the bridge file, then you are not ready to use it in the production releases of your ontology.","title":"1. Add your roots to the OBO bridge"},{"location":"using-cob/#2-use-a-cob-import","text":"Here we assume you are familiar with standard OBO tools such as ROBOT and the ROBOT extract command, or you are using the ODK to make releases. We assume here that all of your imports are to modules extracted from Base files. If you are not doing this, or don't know what a Base file is, then we recommend waiting until the documentation around these is more mature before you attempt to switch to COB. If you are already using a BFO import, then you should modify your build process to instead use COB. If you are configuring an ontology for the first time using the ODK, then all you need to is declare cob as one of your input ontologies. You should do this in place of bfo-classes (don't worry, you will still get most back via COB).","title":"2. Use a COB import"},{"location":"using-cob/#troubleshooting","text":"","title":"Troubleshooting"},{"location":"using-cob/#dangling-bfo-class-ids","text":"The most likely problem will be dangling BFO class IDs. These may come from two sources: subClassOf axioms in a base module from root nodes domain, range, or similar constraining axioms in RO The first case should be rare, because most ontologies are rooted in a COB class with a BFO equivalent. However, in some cases, ontologies will be rooted in either: a class in BFO that is beneath the shoreline in COB, e.g. fiat object part is used as a root in ENVO (for now; see https://github.com/EnvironmentOntology/envo/pull/1252) a class in BFO that is above the shoreline in COB, e.g. a domain ontology class that directly subclasses a high level BFO class like continuant. These should be rare to non-existent The second category is harder. See https://github.com/OBOFoundry/COB/issues/213","title":"Dangling BFO class IDs"},{"location":"using-cob/#using-cob-for-end-users","text":"If you are not an ontology developer or directly involved in the construction of one more ontologies, then there is not much opportunity to \"use\" COB.","title":"Using COB for end-users"},{"location":"odk-workflows/","text":"Default ODK Workflows Daily Editors Workflow Release Workflow Manage your ODK Repository Setting up Docker for ODK Imports management Managing the documentation Managing your Automated Testing","title":"Overview"},{"location":"odk-workflows/#default-odk-workflows","text":"Daily Editors Workflow Release Workflow Manage your ODK Repository Setting up Docker for ODK Imports management Managing the documentation Managing your Automated Testing","title":"Default ODK Workflows"},{"location":"odk-workflows/ContinuousIntegration/","text":"Introduction to Continuous Integration Workflows with ODK Historically, most repos have been using Travis CI for continuous integration testing and building, but due to runtime restrictions, we recently switched a lot of our repos to GitHub actions. You can set up your repo with CI by adding this to your configuration file (src/ontology/cob-odk.yaml): ci: - github_actions When updateing your repo , you will notice a new file being added: .github/workflows/qc.yml . This file contains your CI logic, so if you need to change, or add anything, this is the place! Alternatively, if your repo is in GitLab instead of GitHub, you can set up your repo with GitLab CI by adding this to your configuration file (src/ontology/cob-odk.yaml): ci: - gitlab-ci This will add a file called .gitlab-ci.yml in the root of your repo.","title":"Continuous Integration"},{"location":"odk-workflows/ContinuousIntegration/#introduction-to-continuous-integration-workflows-with-odk","text":"Historically, most repos have been using Travis CI for continuous integration testing and building, but due to runtime restrictions, we recently switched a lot of our repos to GitHub actions. You can set up your repo with CI by adding this to your configuration file (src/ontology/cob-odk.yaml): ci: - github_actions When updateing your repo , you will notice a new file being added: .github/workflows/qc.yml . This file contains your CI logic, so if you need to change, or add anything, this is the place! Alternatively, if your repo is in GitLab instead of GitHub, you can set up your repo with GitLab CI by adding this to your configuration file (src/ontology/cob-odk.yaml): ci: - gitlab-ci This will add a file called .gitlab-ci.yml in the root of your repo.","title":"Introduction to Continuous Integration Workflows with ODK"},{"location":"odk-workflows/EditorsWorkflow/","text":"Editors Workflow The editors workflow is one of the formal workflows to ensure that the ontology is developed correctly according to ontology engineering principles. There are a few different editors workflows: Local editing workflow: Editing the ontology in your local environment by hand, using tools such as Prot\u00e9g\u00e9, ROBOT templates or DOSDP patterns. Completely automated data pipeline (GitHub Actions) DROID workflow This document only covers the first editing workflow, but more will be added in the future Local editing workflow Workflow requirements: git github docker editing tool of choice, e.g. Prot\u00e9g\u00e9, your favourite text editor, etc 1. Create issue Ensure that there is a ticket on your issue tracker that describes the change you are about to make. While this seems optional, this is a very important part of the social contract of building an ontology - no change to the ontology should be performed without a good ticket, describing the motivation and nature of the intended change. 2. Update main branch In your local environment (e.g. your laptop), make sure you are on the main (prev. master ) branch and ensure that you have all the upstream changes, for example: git checkout master git pull 3. Create feature branch Create a new branch. Per convention, we try to use meaningful branch names such as: - issue23removeprocess (where issue 23 is the related issue on GitHub) - issue26addcontributor - release20210101 (for releases) On your command line, this looks like this: git checkout -b issue23removeprocess 4. Perform edit Using your editor of choice, perform the intended edit. For example: Prot\u00e9g\u00e9 Open src/ontology/cob-edit.owl in Prot\u00e9g\u00e9 Make the change Save the file TextEdit Open src/ontology/cob-edit.owl in TextEdit (or Sublime, Atom, Vim, Nano) Make the change Save the file Consider the following when making the edit. According to our development philosophy, the only places that should be manually edited are: src/ontology/cob-edit.owl Any ROBOT templates you chose to use (the TSV files only) Any DOSDP data tables you chose to use (the TSV files, and potentially the associated patterns) components (anything in src/ontology/components ), see here . Imports should not be edited (any edits will be flushed out with the next update). However, refreshing imports is a potentially breaking change - and is discussed elsewhere . Changes should usually be small. Adding or changing 1 term is great. Adding or changing 10 related terms is ok. Adding or changing 100 or more terms at once should be considered very carefully. 4. Check the Git diff This step is very important. Rather than simply trusting your change had the intended effect, we should always use a git diff as a first pass for sanity checking. In our experience, having a visual git client like GitHub Desktop or sourcetree is really helpful for this part. In case you prefer the command line: git status git diff 5. Quality control Now it's time to run your quality control checks. This can either happen locally ( 5a ) or through your continuous integration system ( 7/5b ). 5a. Local testing If you chose to run your test locally: sh run.sh make IMP=false test This will run the whole set of configured ODK tests on including your change. If you have a complex DOSDP pattern pipeline you may want to add PAT=false to skip the potentially lengthy process of rebuilding the patterns. sh run.sh make IMP=false PAT=false test 6. Pull request When you are happy with the changes, you commit your changes to your feature branch, push them upstream (to GitHub) and create a pull request. For example: git add NAMEOFCHANGEDFILES git commit -m \"Added biological process term #12\" git push -u origin issue23removeprocess Then you go to your project on GitHub, and create a new pull request from the branch, for example: https://github.com/INCATools/ontology-development-kit/pulls There is a lot of great advise on how to write pull requests, but at the very least you should: - mention the tickets affected: see #23 to link to a related ticket, or fixes #23 if, by merging this pull request, the ticket is fixed. Tickets in the latter case will be closed automatically by GitHub when the pull request is merged. - summarise the changes in a few sentences. Consider the reviewer: what would they want to know right away. - If the diff is large, provide instructions on how to review the pull request best (sometimes, there are many changed files, but only one important change). 7/5b. Continuous Integration Testing If you didn't run and local quality control checks (see 5a ), you should have Continuous Integration (CI) set up, for example: - Travis - GitHub Actions More on how to set this up here . Once the pull request is created, the CI will automatically trigger. If all is fine, it will show up green, otherwise red. 8. Community review Once all the automatic tests have passed, it is important to put a second set of eyes on the pull request. Ontologies are inherently social - as in that they represent some kind of community consensus on how a domain is organised conceptually. This seems high brow talk, but it is very important that as an ontology editor, you have your work validated by the community you are trying to serve (e.g. your colleagues, other contributors etc.). In our experience, it is hard to get more than one review on a pull request - two is great. You can set up GitHub branch protection to actually require a review before a pull request can be merged! We recommend this. This step seems daunting to some hopefully under-resourced ontologies, but we recommend to put this high up on your list of priorities - train a colleague, reach out! 9. Merge and cleanup When the QC is green and the reviews are in (approvals), it is time to merge the pull request. After the pull request is merged, remember to delete the branch as well (this option will show up as a big button right after you have merged the pull request). If you have not done so, close all the associated tickets fixed by the pull request. 10. Changelog (Optional) It is sometimes difficult to keep track of changes made to an ontology. Some ontology teams opt to document changes in a changelog (simply a text file in your repository) so that when release day comes, you know everything you have changed. This is advisable at least for major changes (such as a new release system, a new pattern or template etc.).","title":"Editors Workflow"},{"location":"odk-workflows/EditorsWorkflow/#editors-workflow","text":"The editors workflow is one of the formal workflows to ensure that the ontology is developed correctly according to ontology engineering principles. There are a few different editors workflows: Local editing workflow: Editing the ontology in your local environment by hand, using tools such as Prot\u00e9g\u00e9, ROBOT templates or DOSDP patterns. Completely automated data pipeline (GitHub Actions) DROID workflow This document only covers the first editing workflow, but more will be added in the future","title":"Editors Workflow"},{"location":"odk-workflows/EditorsWorkflow/#local-editing-workflow","text":"Workflow requirements: git github docker editing tool of choice, e.g. Prot\u00e9g\u00e9, your favourite text editor, etc","title":"Local editing workflow"},{"location":"odk-workflows/EditorsWorkflow/#1-create-issue","text":"Ensure that there is a ticket on your issue tracker that describes the change you are about to make. While this seems optional, this is a very important part of the social contract of building an ontology - no change to the ontology should be performed without a good ticket, describing the motivation and nature of the intended change.","title":"1. Create issue"},{"location":"odk-workflows/EditorsWorkflow/#2-update-main-branch","text":"In your local environment (e.g. your laptop), make sure you are on the main (prev. master ) branch and ensure that you have all the upstream changes, for example: git checkout master git pull","title":"2. Update main branch"},{"location":"odk-workflows/EditorsWorkflow/#3-create-feature-branch","text":"Create a new branch. Per convention, we try to use meaningful branch names such as: - issue23removeprocess (where issue 23 is the related issue on GitHub) - issue26addcontributor - release20210101 (for releases) On your command line, this looks like this: git checkout -b issue23removeprocess","title":"3. Create feature branch"},{"location":"odk-workflows/EditorsWorkflow/#4-perform-edit","text":"Using your editor of choice, perform the intended edit. For example: Prot\u00e9g\u00e9 Open src/ontology/cob-edit.owl in Prot\u00e9g\u00e9 Make the change Save the file TextEdit Open src/ontology/cob-edit.owl in TextEdit (or Sublime, Atom, Vim, Nano) Make the change Save the file Consider the following when making the edit. According to our development philosophy, the only places that should be manually edited are: src/ontology/cob-edit.owl Any ROBOT templates you chose to use (the TSV files only) Any DOSDP data tables you chose to use (the TSV files, and potentially the associated patterns) components (anything in src/ontology/components ), see here . Imports should not be edited (any edits will be flushed out with the next update). However, refreshing imports is a potentially breaking change - and is discussed elsewhere . Changes should usually be small. Adding or changing 1 term is great. Adding or changing 10 related terms is ok. Adding or changing 100 or more terms at once should be considered very carefully.","title":"4. Perform edit"},{"location":"odk-workflows/EditorsWorkflow/#4-check-the-git-diff","text":"This step is very important. Rather than simply trusting your change had the intended effect, we should always use a git diff as a first pass for sanity checking. In our experience, having a visual git client like GitHub Desktop or sourcetree is really helpful for this part. In case you prefer the command line: git status git diff","title":"4. Check the Git diff"},{"location":"odk-workflows/EditorsWorkflow/#5-quality-control","text":"Now it's time to run your quality control checks. This can either happen locally ( 5a ) or through your continuous integration system ( 7/5b ).","title":"5. Quality control"},{"location":"odk-workflows/EditorsWorkflow/#5a-local-testing","text":"If you chose to run your test locally: sh run.sh make IMP=false test This will run the whole set of configured ODK tests on including your change. If you have a complex DOSDP pattern pipeline you may want to add PAT=false to skip the potentially lengthy process of rebuilding the patterns. sh run.sh make IMP=false PAT=false test","title":"5a. Local testing"},{"location":"odk-workflows/EditorsWorkflow/#6-pull-request","text":"When you are happy with the changes, you commit your changes to your feature branch, push them upstream (to GitHub) and create a pull request. For example: git add NAMEOFCHANGEDFILES git commit -m \"Added biological process term #12\" git push -u origin issue23removeprocess Then you go to your project on GitHub, and create a new pull request from the branch, for example: https://github.com/INCATools/ontology-development-kit/pulls There is a lot of great advise on how to write pull requests, but at the very least you should: - mention the tickets affected: see #23 to link to a related ticket, or fixes #23 if, by merging this pull request, the ticket is fixed. Tickets in the latter case will be closed automatically by GitHub when the pull request is merged. - summarise the changes in a few sentences. Consider the reviewer: what would they want to know right away. - If the diff is large, provide instructions on how to review the pull request best (sometimes, there are many changed files, but only one important change).","title":"6. Pull request"},{"location":"odk-workflows/EditorsWorkflow/#75b-continuous-integration-testing","text":"If you didn't run and local quality control checks (see 5a ), you should have Continuous Integration (CI) set up, for example: - Travis - GitHub Actions More on how to set this up here . Once the pull request is created, the CI will automatically trigger. If all is fine, it will show up green, otherwise red.","title":"7/5b. Continuous Integration Testing"},{"location":"odk-workflows/EditorsWorkflow/#8-community-review","text":"Once all the automatic tests have passed, it is important to put a second set of eyes on the pull request. Ontologies are inherently social - as in that they represent some kind of community consensus on how a domain is organised conceptually. This seems high brow talk, but it is very important that as an ontology editor, you have your work validated by the community you are trying to serve (e.g. your colleagues, other contributors etc.). In our experience, it is hard to get more than one review on a pull request - two is great. You can set up GitHub branch protection to actually require a review before a pull request can be merged! We recommend this. This step seems daunting to some hopefully under-resourced ontologies, but we recommend to put this high up on your list of priorities - train a colleague, reach out!","title":"8. Community review"},{"location":"odk-workflows/EditorsWorkflow/#9-merge-and-cleanup","text":"When the QC is green and the reviews are in (approvals), it is time to merge the pull request. After the pull request is merged, remember to delete the branch as well (this option will show up as a big button right after you have merged the pull request). If you have not done so, close all the associated tickets fixed by the pull request.","title":"9. Merge and cleanup"},{"location":"odk-workflows/EditorsWorkflow/#10-changelog-optional","text":"It is sometimes difficult to keep track of changes made to an ontology. Some ontology teams opt to document changes in a changelog (simply a text file in your repository) so that when release day comes, you know everything you have changed. This is advisable at least for major changes (such as a new release system, a new pattern or template etc.).","title":"10. Changelog (Optional)"},{"location":"odk-workflows/ManageDocumentation/","text":"Updating the Documentation The documentation for COB is managed in two places (relative to the repository root): The docs directory contains all the files that pertain to the content of the documentation (more below) the mkdocs.yaml file contains the documentation config, in particular its navigation bar and theme. The documentation is hosted using GitHub pages, on a special branch of the repository (called gh-pages ). It is important that this branch is never deleted - it contains all the files GitHub pages needs to render and deploy the site. It is also important to note that the gh-pages branch should never be edited manually . All changes to the docs happen inside the docs directory on the main branch. Editing the docs Changing content All the documentation is contained in the docs directory, and is managed in Markdown . Markdown is a very simple and convenient way to produce text documents with formatting instructions, and is very easy to learn - it is also used, for example, in GitHub issues. This is a normal editing workflow: Open the .md file you want to change in an editor of choice (a simple text editor is often best). IMPORTANT : Do not edit any files in the docs/odk-workflows/ directory. These files are managed by the ODK system and will be overwritten when the repository is upgraded! If you wish to change these files, make an issue on the ODK issue tracker . Perform the edit and save the file Commit the file to a branch, and create a pull request as usual. If your development team likes your changes, merge the docs into master branch. Deploy the documentation (see below) Deploy the documentation The documentation is not automatically updated from the Markdown, and needs to be deployed deliberately. To do this, perform the following steps: In your terminal, navigate to the edit directory of your ontology, e.g.: cd cob/src/ontology Now you are ready to build the docs as follows: sh run.sh make update_docs Mkdocs now sets off to build the site from the markdown pages. You will be asked to Enter your username Enter your password (see here for using GitHub access tokens instead) IMPORTANT : Using password based authentication will be deprecated this year (2021). Make sure you read up on personal access tokens if that happens! If everything was successful, you will see a message similar to this one: INFO - Your documentation should shortly be available at: https://OBOFoundry.github.io/COB/ 3. Just to double check, you can now navigate to your documentation pages (usually https://OBOFoundry.github.io/COB/). Just make sure you give GitHub 2-5 minutes to build the pages!","title":"Managing the documentation"},{"location":"odk-workflows/ManageDocumentation/#updating-the-documentation","text":"The documentation for COB is managed in two places (relative to the repository root): The docs directory contains all the files that pertain to the content of the documentation (more below) the mkdocs.yaml file contains the documentation config, in particular its navigation bar and theme. The documentation is hosted using GitHub pages, on a special branch of the repository (called gh-pages ). It is important that this branch is never deleted - it contains all the files GitHub pages needs to render and deploy the site. It is also important to note that the gh-pages branch should never be edited manually . All changes to the docs happen inside the docs directory on the main branch.","title":"Updating the Documentation"},{"location":"odk-workflows/ManageDocumentation/#editing-the-docs","text":"","title":"Editing the docs"},{"location":"odk-workflows/ManageDocumentation/#changing-content","text":"All the documentation is contained in the docs directory, and is managed in Markdown . Markdown is a very simple and convenient way to produce text documents with formatting instructions, and is very easy to learn - it is also used, for example, in GitHub issues. This is a normal editing workflow: Open the .md file you want to change in an editor of choice (a simple text editor is often best). IMPORTANT : Do not edit any files in the docs/odk-workflows/ directory. These files are managed by the ODK system and will be overwritten when the repository is upgraded! If you wish to change these files, make an issue on the ODK issue tracker . Perform the edit and save the file Commit the file to a branch, and create a pull request as usual. If your development team likes your changes, merge the docs into master branch. Deploy the documentation (see below)","title":"Changing content"},{"location":"odk-workflows/ManageDocumentation/#deploy-the-documentation","text":"The documentation is not automatically updated from the Markdown, and needs to be deployed deliberately. To do this, perform the following steps: In your terminal, navigate to the edit directory of your ontology, e.g.: cd cob/src/ontology Now you are ready to build the docs as follows: sh run.sh make update_docs Mkdocs now sets off to build the site from the markdown pages. You will be asked to Enter your username Enter your password (see here for using GitHub access tokens instead) IMPORTANT : Using password based authentication will be deprecated this year (2021). Make sure you read up on personal access tokens if that happens! If everything was successful, you will see a message similar to this one: INFO - Your documentation should shortly be available at: https://OBOFoundry.github.io/COB/ 3. Just to double check, you can now navigate to your documentation pages (usually https://OBOFoundry.github.io/COB/). Just make sure you give GitHub 2-5 minutes to build the pages!","title":"Deploy the documentation"},{"location":"odk-workflows/ReleaseWorkflow/","text":"The release workflow The release workflow recommended by the ODK is based on GitHub releases and works as follows: Run a release with the ODK Review the release Merge to main branch Create a GitHub release These steps are outlined in detail in the following. Run a release with the ODK Preparation: Ensure that all your pull requests are merged into your main (master) branch Make sure that all changes to master are committed to GitHub ( git status should say that there are no modified files) Locally make sure you have the latest changes from master ( git pull ) Checkout a new branch (e.g. git checkout -b release-2021-01-01 ) You may or may not want to refresh your imports as part of your release strategy (see here ) Make sure you have the latest ODK installed by running docker pull obolibrary/odkfull To actually run the release, you: Open a command line terminal window and navigate to the src/ontology directory ( cd cob/src/ontology ) Run release pipeline: sh run.sh make prepare_release -B . Note that for some ontologies, this process can take up to 90 minutes - especially if there are large ontologies you depend on, like PRO or CHEBI. If everything went well, you should see the following output on your machine: Release files are now in ../.. - now you should commit, push and make a release on your git hosting site such as GitHub or GitLab . This will create all the specified release targets (OBO, OWL, JSON, and the variants, ont-full and ont-base) and copy them into your release directory (the top level of your repo). Review the release (Optional) Rough check. This step is frequently skipped, but for the more paranoid among us (like the author of this doc), this is a 3 minute additional effort for some peace of mind. Open the main release (cob.owl) in you favourite development environment (i.e. Prot\u00e9g\u00e9) and eyeball the hierarchy. We recommend two simple checks: Does the very top level of the hierarchy look ok? This means that all new terms have been imported/updated correctly. Does at least one change that you know should be in this release appear? For example, a new class. This means that the release was actually based on the recent edit file. Commit your changes to the branch and make a pull request In your GitHub pull request, review the following three files in detail (based on our experience): cob.obo - this reflects a useful subset of the whole ontology (everything that can be covered by OBO format). OBO format has that speaking for it: it is very easy to review! cob-base.owl - this reflects the asserted axioms in your ontology that you have actually edited. Ideally also take a look at cob-full.owl , which may reveal interesting new inferences you did not know about. Note that the diff of this file is sometimes quite large. Like with every pull request, we recommend to always employ a second set of eyes when reviewing a PR! Merge the main branch Once your CI checks have passed, and your reviews are completed, you can now merge the branch into your main branch (don't forget to delete the branch afterwards - a big button will appear after the merge is finished). Create a GitHub release Go to your releases page on GitHub by navigating to your repository, and then clicking on releases (usually on the right, for example: https://github.com/OBOFoundry/COB/releases). Then click \"Draft new release\" As the tag version you need to choose the date on which your ontologies were build. You can find this, for example, by looking at the cob.obo file and check the data-version: property. The date needs to be prefixed with a v , so, for example v2020-02-06 . You can write whatever you want in the release title, but we typically write the date again. The description underneath should contain a concise list of changes or term additions. Click \"Publish release\". Done. Debugging typical ontology release problems Problems with memory When you are dealing with large ontologies, you need a lot of memory. When you see error messages relating to large ontologies such as CHEBI, PRO, NCBITAXON, or Uberon, you should think of memory first, see here . Problems when using OBO format based tools Sometimes you will get cryptic error messages when using legacy tools using OBO format, such as the ontology release tool (OORT), which is also available as part of the ODK docker container. In these cases, you need to track down what axiom or annotation actually caused the breakdown. In our experience (in about 60% of the cases) the problem lies with duplicate annotations ( def , comment ) which are illegal in OBO. Here is an example recipe of how to deal with such a problem: If you get a message like make: *** [cl.Makefile:84: oort] Error 255 you might have a OORT error. To debug this, in your terminal enter sh run.sh make IMP=false PAT=false oort -B (assuming you are already in the ontology folder in your directory) This should show you where the error is in the log (eg multiple different definitions) WARNING: THE FIX BELOW IS NOT IDEAL, YOU SHOULD ALWAYS TRY TO FIX UPSTREAM IF POSSIBLE Open cob-edit.owl in Prot\u00e9g\u00e9 and find the offending term and delete all offending issue (e.g. delete ALL definition, if the problem was \"multiple def tags not allowed\") and save. *While this is not idea, as it will remove all definitions from that term, it will be added back again when the term is fixed in the ontology it was imported from and added back in. Rerun sh run.sh make IMP=false PAT=false oort -B and if it all passes, commit your changes to a branch and make a pull request as usual.","title":"Release Workflow"},{"location":"odk-workflows/ReleaseWorkflow/#the-release-workflow","text":"The release workflow recommended by the ODK is based on GitHub releases and works as follows: Run a release with the ODK Review the release Merge to main branch Create a GitHub release These steps are outlined in detail in the following.","title":"The release workflow"},{"location":"odk-workflows/ReleaseWorkflow/#run-a-release-with-the-odk","text":"Preparation: Ensure that all your pull requests are merged into your main (master) branch Make sure that all changes to master are committed to GitHub ( git status should say that there are no modified files) Locally make sure you have the latest changes from master ( git pull ) Checkout a new branch (e.g. git checkout -b release-2021-01-01 ) You may or may not want to refresh your imports as part of your release strategy (see here ) Make sure you have the latest ODK installed by running docker pull obolibrary/odkfull To actually run the release, you: Open a command line terminal window and navigate to the src/ontology directory ( cd cob/src/ontology ) Run release pipeline: sh run.sh make prepare_release -B . Note that for some ontologies, this process can take up to 90 minutes - especially if there are large ontologies you depend on, like PRO or CHEBI. If everything went well, you should see the following output on your machine: Release files are now in ../.. - now you should commit, push and make a release on your git hosting site such as GitHub or GitLab . This will create all the specified release targets (OBO, OWL, JSON, and the variants, ont-full and ont-base) and copy them into your release directory (the top level of your repo).","title":"Run a release with the ODK"},{"location":"odk-workflows/ReleaseWorkflow/#review-the-release","text":"(Optional) Rough check. This step is frequently skipped, but for the more paranoid among us (like the author of this doc), this is a 3 minute additional effort for some peace of mind. Open the main release (cob.owl) in you favourite development environment (i.e. Prot\u00e9g\u00e9) and eyeball the hierarchy. We recommend two simple checks: Does the very top level of the hierarchy look ok? This means that all new terms have been imported/updated correctly. Does at least one change that you know should be in this release appear? For example, a new class. This means that the release was actually based on the recent edit file. Commit your changes to the branch and make a pull request In your GitHub pull request, review the following three files in detail (based on our experience): cob.obo - this reflects a useful subset of the whole ontology (everything that can be covered by OBO format). OBO format has that speaking for it: it is very easy to review! cob-base.owl - this reflects the asserted axioms in your ontology that you have actually edited. Ideally also take a look at cob-full.owl , which may reveal interesting new inferences you did not know about. Note that the diff of this file is sometimes quite large. Like with every pull request, we recommend to always employ a second set of eyes when reviewing a PR!","title":"Review the release"},{"location":"odk-workflows/ReleaseWorkflow/#merge-the-main-branch","text":"Once your CI checks have passed, and your reviews are completed, you can now merge the branch into your main branch (don't forget to delete the branch afterwards - a big button will appear after the merge is finished).","title":"Merge the main branch"},{"location":"odk-workflows/ReleaseWorkflow/#create-a-github-release","text":"Go to your releases page on GitHub by navigating to your repository, and then clicking on releases (usually on the right, for example: https://github.com/OBOFoundry/COB/releases). Then click \"Draft new release\" As the tag version you need to choose the date on which your ontologies were build. You can find this, for example, by looking at the cob.obo file and check the data-version: property. The date needs to be prefixed with a v , so, for example v2020-02-06 . You can write whatever you want in the release title, but we typically write the date again. The description underneath should contain a concise list of changes or term additions. Click \"Publish release\". Done.","title":"Create a GitHub release"},{"location":"odk-workflows/ReleaseWorkflow/#debugging-typical-ontology-release-problems","text":"","title":"Debugging typical ontology release problems"},{"location":"odk-workflows/ReleaseWorkflow/#problems-with-memory","text":"When you are dealing with large ontologies, you need a lot of memory. When you see error messages relating to large ontologies such as CHEBI, PRO, NCBITAXON, or Uberon, you should think of memory first, see here .","title":"Problems with memory"},{"location":"odk-workflows/ReleaseWorkflow/#problems-when-using-obo-format-based-tools","text":"Sometimes you will get cryptic error messages when using legacy tools using OBO format, such as the ontology release tool (OORT), which is also available as part of the ODK docker container. In these cases, you need to track down what axiom or annotation actually caused the breakdown. In our experience (in about 60% of the cases) the problem lies with duplicate annotations ( def , comment ) which are illegal in OBO. Here is an example recipe of how to deal with such a problem: If you get a message like make: *** [cl.Makefile:84: oort] Error 255 you might have a OORT error. To debug this, in your terminal enter sh run.sh make IMP=false PAT=false oort -B (assuming you are already in the ontology folder in your directory) This should show you where the error is in the log (eg multiple different definitions) WARNING: THE FIX BELOW IS NOT IDEAL, YOU SHOULD ALWAYS TRY TO FIX UPSTREAM IF POSSIBLE Open cob-edit.owl in Prot\u00e9g\u00e9 and find the offending term and delete all offending issue (e.g. delete ALL definition, if the problem was \"multiple def tags not allowed\") and save. *While this is not idea, as it will remove all definitions from that term, it will be added back again when the term is fixed in the ontology it was imported from and added back in. Rerun sh run.sh make IMP=false PAT=false oort -B and if it all passes, commit your changes to a branch and make a pull request as usual.","title":"Problems when using OBO format based tools"},{"location":"odk-workflows/RepoManagement/","text":"Managing your ODK repository Updating your ODK repository Your ODK repositories configuration is managed in src/ontology/cob-odk.yaml . Once you have made your changes, you can run the following to apply your changes to the repository: sh run.sh make update_repo There are a large number of options that can be set to configure your ODK, but we will only discuss a few of them here. NOTE for Windows users: You may get a cryptic failure such as Set Illegal Option - if the update script located in src/scripts/update_repo.sh was saved using Windows Line endings. These need to change to unix line endings. In Notepad++, for example, you can click on Edit->EOL Conversion->Unix LF to change this. Managing imports You can use the update repository workflow described on this page to perform the following operations to your imports: Add a new import Modify an existing import Remove an import you no longer want Customise an import We will discuss all these workflows in the following. Add new import To add a new import, you first edit your odk config as described above , adding an id to the product list in the import_group section (for the sake of this example, we assume you already import RO, and your goal is to also import GO): import_group: products: - id: ro - id: go Note: our ODK file should only have one import_group which can contain multiple imports (in the products section). Next, you run the update repo workflow to apply these changes. Note that by default, this module is going to be a SLME Bottom module, see here . To change that or customise your module, see section \"Customise an import\". To finalise the addition of your import, perform the following steps: Add an import statement to your src/ontology/cob-edit.owl file. We suggest to do this using a text editor, by simply copying an existing import declaration and renaming it to the new ontology import, for example as follows: ... Ontology(Using COB: An ontology developers guide
+Intended audience: technical leads for individual OBO ontology projects
+This is a draft guide. There are many technical issues that need to be +worked out to transition to COB. We strongly recommend you do work +here on a branch, and if you have problems, make an issue in the COB +tracker and link to your PR
+1. Add your roots to the OBO bridge
+See Bridging COB to OBO for instructions on how to make sure your ontology's root terms are mapped to COB.
+If you are not bridged to COB, or don't understand the bridge file, then you are not ready to use it in the production releases of your ontology.
+2. Use a COB import
+Here we assume you are familiar with standard OBO tools such as ROBOT and the ROBOT extract command, or you are using the ODK to make releases.
+We assume here that all of your imports are to modules extracted from Base files. If you are not doing this, or don't know what a Base file is, then we recommend waiting until the documentation around these is more mature before you attempt to switch to COB.
+If you are already using a BFO import, then you should modify your build process to instead use COB.
+If you are configuring an ontology for the first time using the ODK, then all you need to is declare cob as one of your input ontologies. You should do this in place of bfo-classes (don't worry, you will still get most back via COB).
Troubleshooting
+Dangling BFO class IDs
+The most likely problem will be dangling BFO class IDs. These may come from two sources:
+-
+
- subClassOf axioms in a base module from root nodes +
- domain, range, or similar constraining axioms in RO +
The first case should be rare, because most ontologies are rooted in a COB class with a BFO equivalent. However, in some cases, ontologies will be rooted in either:
+-
+
- a class in BFO that is beneath the shoreline in COB, e.g. fiat object part is used as a root in ENVO (for now; see https://github.com/EnvironmentOntology/envo/pull/1252) +
- a class in BFO that is above the shoreline in COB, e.g. a domain ontology class that directly subclasses a high level BFO class like continuant. These should be rare to non-existent +
The second category is harder. See https://github.com/OBOFoundry/COB/issues/213
+Using COB for end-users
+If you are not an ontology developer or directly involved in the construction of one more ontologies, then there is not much opportunity to "use" COB.
+ + + + + + +