-
- En svensk översättning av denna sida kommer inom kort.
-
-
-The visualisations on this page evaluate the development of COVID-19 and SARS-CoV-2 research across Sweden by assessing publication output. Specifically, we consider multiple aspects of journal publications and preprints where at least one author has an affiliation with a Swedish research institute. The database containing the publications themselves [can be found on this page](/publications/), and is available for download, please see [DOI: 10.17044/scilifelab.14124014](https://doi.org/10.17044/scilifelab.14124014) for details. The database is manually curated, so may not be exhaustive. From May 2023, we began to use the Europe PMC REST API to idenfy publications. The scripts that we use to do this are [openly available on GitHub](https://github.com/ScilifelabDataCentre/pathogens-portal-scripts/tree/main/All_publications) and can be reused for work with other pathogens. The database is updated monthly at the start of the month.
-
-The code used to produce the visulations on this page can be found on [GitHub](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations). Specifically, code related to the number of publications can be found in the ['Count_publications' folder of the repository](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Count_publications), and code used to generated the wordclouds can be found in the ['Wordcloud' folder](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Wordcloud).
+Visualiseringarna på denna sida återspeglar hur produktionen av vetenskapliga forskningsartiklar kopplade till covid-19 och SARS-CoV-2 utvecklas över tid. Vi har valt att presentera en sammanställning av publicerade vetenskapliga artiklar och preprint där minst en författare är affilierad till ett svenskt lärosäte. Den svenska covid-19 publikationsdatabasen [nås här](/sv/publications/). Observera att sammanställningen görs manuellt och kan därför inte kan anses komplett. Den fullständiga sammanställningen av publikationer som visas på denna sida finns tillgänglig för nedladdning och användning för andra ändamål, se [DOI: 10.17044/scilifelab.14124014](https://doi.org/10.17044/scilifelab.14124014). Databasen är manuellt kuraterad, så den kan vara ofullständig. Från och med maj 2023 började vi använda Europe PMC REST API för att identifiera publikationer. Skripten som vi använder för detta är [öppet tillgängliga på GitHub](https://github.com/ScilifelabDataCentre/pathogens-portal-scripts/tree/main/All_publications) och kan återanvändas för arbete med andra patogener. Databasen uppdateras månadsvis i början av månaden.
+Koden som används för att producera visualiseringarna på denna sida finns på [GitHub](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations). Specifikt kan kod relaterad till antalet publikationer hittas i ['Count_publications' mappen i förvaret](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Count_publications), och kod som används för att generera ordmolnen kan hittas i ['Wordcloud' mappen](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Wordcloud).
{{% publication_updated_date %}}
-## Number of new publications
+## Antal nya publikationer
-This graph displays the number of publications (including both journal publications and preprints) published each month, as well as the cumulative daily total of publications contained in the database. The dates reflect either the dates that the articles were uploaded to preprint servers (in the case of preprints) or the official journal publication date, whichever is the most recent. Where a given day of publication is not specified, we assign the date as the first of the month. This causes the appearance of a relatively sharp increase at the start of each month.
+Diagrammet visar antal publikationer (vilket inkluderar vetenskapliga artiklar och preprint) som publiceras varje månad samt det sammanlagda antalet publikationer per dag. Datumen återspeglar uppladdningsdatumet för preprint eller officiellt publuceringsdatumet för vetenskapliga artiklar, beroende på vilket som är det senaste. Att antalet publikationer tycks vara högt första dagen i varje månad beror på att inte alla publikationer har ett publikationsdatum; alla publikationer utan specifikt publikationsdatum har lagts in som att de publicerats den första dagen I månaden.
Rotating your phone may improve graph layout
@@ -37,27 +31,23 @@ This graph displays the number of publications (including both journal publicati
-
-
-**Code used to produce plot:** [Script to produce plot](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/blob/main/Count_publications/count_publications.py).
+**Källkod som används för att skapa grafen:** [Källkod som används för att skapa visualisering](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/blob/main/Count_publications/count_publications.py).
-## Ten most recent publications
+## De senast publicerade publikationerna (tio senaste)
-This table displays the ten most recent publications in our database (including both journal publications and preprints). The date reflects the preprint upload date or the official journal publication date, whichever is the most recent. Authors are summarised in the same format as expected in 'in text' citations. Clicking on the title for a given publication will take you directly to that publication. Note though, that it will only be possible to read the full text of publication if it is open access or if you are granted access via a subscription (e.g. an institutional subscription, or private journal subscripton). Any additions to the manually curated publications database will become visible on this table within a day.
+Tabellen visar de tio senaste publikationerna (inkluderar både tidskriftsartiklar och preprints). Datumet som anges är antingen det första datumet ett preprint laddades upp på en preprintserver eller publikationsdatumet för vetenskapliga artiklar. Författarlistan anges i samma format som citeringar. Genom att klicka på en specifik publikation länkas du vidare till publikationen. Observera att det endast är möjligt att läsa fulltext versionen av publikationer som publiceras open access eller ifall du har tillgång till en prenumeration via högskola eller universitet samt via en egen tidskriftsprenumeration. Tillägg till den manuellt granskade publikationsdatabasen kommer bli synliga i denna tabell nästa dag.
{{< plotly json="https://blobserver.dc.scilifelab.se/blob/COVID_publication_count.json" height="600px" >}}
-{{< recent_ten >}}
+{{< recent_ten_swe >}}
-## Most frequent words or two word phrases in the titles
+## Vanligast förekommande ord eller fraser i titlarna
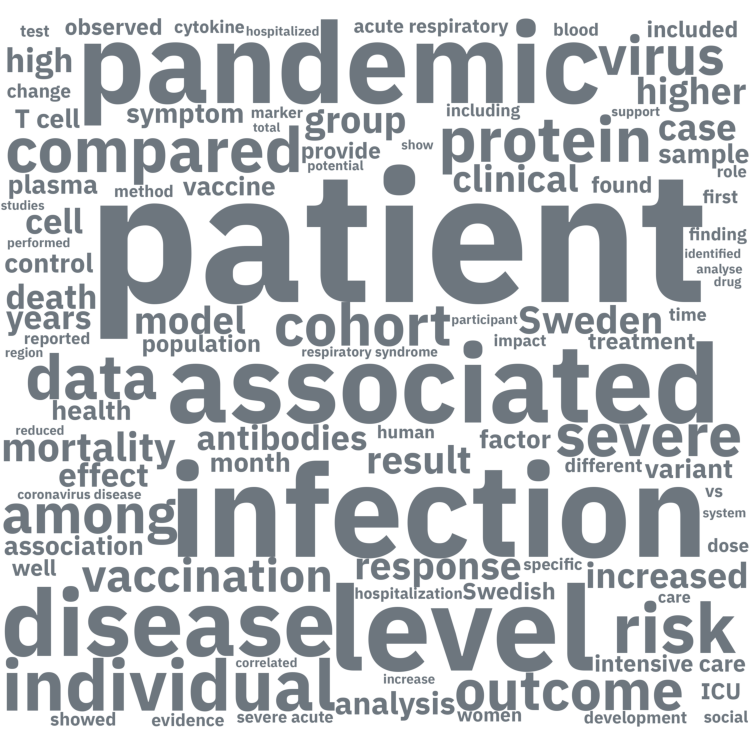
-These wordclouds display the words and two word phrases that appear most frequently in the titles of preprints or journal publications within the database. Note that we have filtered out commonly used words that are uninformative (e.g. 'the', 'a', 'this') as well as the words 'COVID-19' and 'SARS-CoV-2', since these appeared in almost all titles. The wordclouds are updated weekly.
+Detta ordmoln visar de vanligaste orden eller fraserna som förekommer i publikationernas titlar. Observera att vi har filtrerat bort funktionella ord (som 'the', 'a', 'this', etc.) samt 'COVID-19' och 'SARS-CoV-2' eftersom alla titlar innehöll dessa ord. Alla ordmoln uppdateras veckovis.
-**Code used to produce plot:** [Script to produce wordclouds](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/blob/main/Wordcloud/livewordcloud.py). Note that the script relies on multiple external files that can be found in the [same folder in the GitHub repository](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Wordcloud).
+**Källkod som används för att skapa ordmoln:** [Källkod som används för att skapa visualiseringarna](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Wordcloud).
-#### All publications
+#### Alla publikationer
@@ -65,15 +55,15 @@ These wordclouds display the words and two word phrases that appear most frequen
-
Swedish Research Council:
+Vetenskapsrådet:
 @@ -98,13 +88,13 @@ Wordclouds are displayed for each funder that we identified as having been assoc
@@ -98,13 +88,13 @@ Wordclouds are displayed for each funder that we identified as having been assoc
@@ -112,15 +102,15 @@ These wordclouds display the words and two word phrases that appear most frequen
-
Swedish Research Council:
+Vetenskapsrådet:
 diff --git a/content/svenska/projects/_index.md b/content/svenska/projects/_index.md
deleted file mode 100644
index 6bdcd983b..000000000
--- a/content/svenska/projects/_index.md
+++ /dev/null
@@ -1,7 +0,0 @@
----
-title: Forskningsprojekt relaterade till covid-19
----
-
-- [Publikationer rörande covid-19 från svenska lärosäten](/sv/publications/)
-- [Översikt över pågående forskningsprojekt relaterade till covid-19](/sv/projects/ongoing/)
-- [Översikt över kommande forskningsanslag relaterade till covid-19](/sv/projects/funding/)
diff --git a/content/svenska/projects/dashboard.md b/content/svenska/projects/dashboard.md
deleted file mode 100644
index 42241d5c1..000000000
--- a/content/svenska/projects/dashboard.md
+++ /dev/null
@@ -1,58 +0,0 @@
----
-title: Visualisering av svensk covid-19 forskning
-toc: false
-plotly: true
----
-Visualiseringarna på denna sida återspeglar hur produktionen av vetenskapliga forskningsartiklar kopplade till covid-19 och SARS-CoV-2 utvecklas över tid. Vi har valt att presentera en sammanställning av publicerade vetenskapliga artiklar och preprint där minst en författare är affilierad till ett svenskt lärosäte. Den svenska covid-19 publikationsdatabasen [nås här](/sv/publications/). Observera att sammanställningen görs manuellt och kan därför inte kan anses komplett. Den fullständiga sammanställningen av publikationer som visas på denna sida finns tillgänglig för nedladdning och användning för andra ändamål, se [DOI: 10.17044/scilifelab.14124014](https://doi.org/10.17044/scilifelab.14124014).
-
-All källkod som används för att göra visualiseringarna på denna sidan finns tillgängliga på [GitHub](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations). Källkod som används är länkad under respektive visualisering.
-
-## Antal nya publikationer
-
-Diagrammet visar antal publikationer (vilket inkluderar vetenskapliga artiklar och preprint) som publiceras varje månad samt det sammanlagda antalet publikationer per dag. Datumen återspeglar uppladdningsdatumet för preprint eller officiellt publuceringsdatumet för vetenskapliga artiklar, beroende på vilket som är det senaste. Att antalet publikationer tycks vara högt första dagen i varje månad beror på att inte alla publikationer har ett publikationsdatum; alla publikationer utan specifikt publikationsdatum har lagts in som att de publicerats den första dagen I månaden.
-
-
diff --git a/content/svenska/projects/_index.md b/content/svenska/projects/_index.md
deleted file mode 100644
index 6bdcd983b..000000000
--- a/content/svenska/projects/_index.md
+++ /dev/null
@@ -1,7 +0,0 @@
----
-title: Forskningsprojekt relaterade till covid-19
----
-
-- [Publikationer rörande covid-19 från svenska lärosäten](/sv/publications/)
-- [Översikt över pågående forskningsprojekt relaterade till covid-19](/sv/projects/ongoing/)
-- [Översikt över kommande forskningsanslag relaterade till covid-19](/sv/projects/funding/)
diff --git a/content/svenska/projects/dashboard.md b/content/svenska/projects/dashboard.md
deleted file mode 100644
index 42241d5c1..000000000
--- a/content/svenska/projects/dashboard.md
+++ /dev/null
@@ -1,58 +0,0 @@
----
-title: Visualisering av svensk covid-19 forskning
-toc: false
-plotly: true
----
-Visualiseringarna på denna sida återspeglar hur produktionen av vetenskapliga forskningsartiklar kopplade till covid-19 och SARS-CoV-2 utvecklas över tid. Vi har valt att presentera en sammanställning av publicerade vetenskapliga artiklar och preprint där minst en författare är affilierad till ett svenskt lärosäte. Den svenska covid-19 publikationsdatabasen [nås här](/sv/publications/). Observera att sammanställningen görs manuellt och kan därför inte kan anses komplett. Den fullständiga sammanställningen av publikationer som visas på denna sida finns tillgänglig för nedladdning och användning för andra ändamål, se [DOI: 10.17044/scilifelab.14124014](https://doi.org/10.17044/scilifelab.14124014).
-
-All källkod som används för att göra visualiseringarna på denna sidan finns tillgängliga på [GitHub](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations). Källkod som används är länkad under respektive visualisering.
-
-## Antal nya publikationer
-
-Diagrammet visar antal publikationer (vilket inkluderar vetenskapliga artiklar och preprint) som publiceras varje månad samt det sammanlagda antalet publikationer per dag. Datumen återspeglar uppladdningsdatumet för preprint eller officiellt publuceringsdatumet för vetenskapliga artiklar, beroende på vilket som är det senaste. Att antalet publikationer tycks vara högt första dagen i varje månad beror på att inte alla publikationer har ett publikationsdatum; alla publikationer utan specifikt publikationsdatum har lagts in som att de publicerats den första dagen I månaden.
-
-
-{{< publications_per_month >}}
-
-
-**Källkod som används för att skapa grafen:** [Källkod som används för att skapa visualisering](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/blob/main/Count_publications/count_publications.py).
-
-## De senast publicerade publikationerna (tio senaste)
-
-Tabellen visar de tio senaste publikationerna (inkluderar både tidskriftsartiklar och preprints). Datumet som anges är antingen det första datumet ett preprint laddades upp på en preprintserver eller publikationsdatumet för vetenskapliga artiklar. Författarlistan anges i samma format som citeringar. Genom att klicka på en specifik publikation länkas du vidare till publikationen. Observera att det endast är möjligt att läsa fulltext versionen av publikationer som publiceras open access eller ifall du har tillgång till en prenumeration via högskola eller universitet samt via en egen tidskriftsprenumeration. Tillägg till den manuellt granskade publikationsdatabasen kommer bli synliga i denna tabell nästa dag.
-
-
-{{< recent_ten_swe >}}
-
-
-## Vanligast förekommande ord eller fraser i titlarna
-
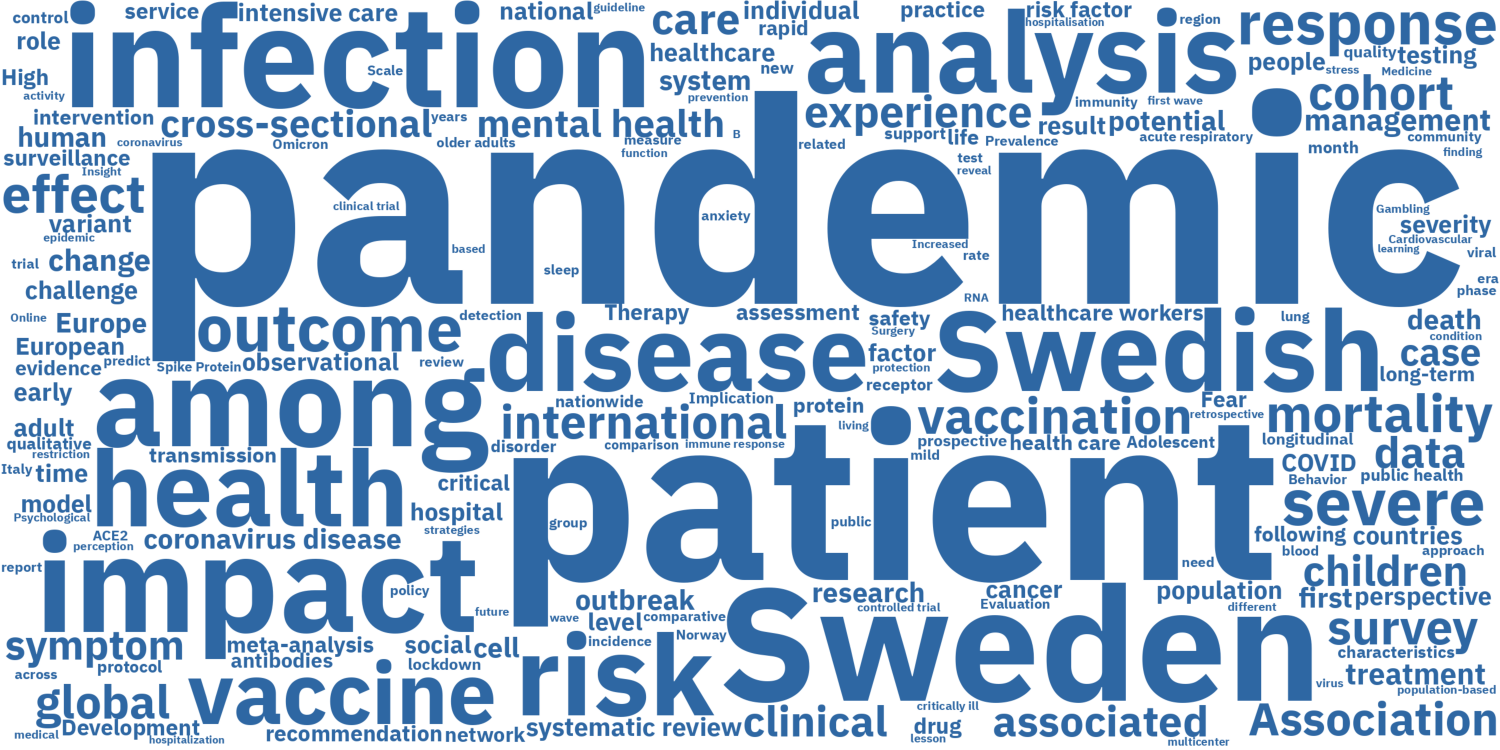
-Detta ordmoln visar de vanligaste orden eller fraserna som förekommer i publikationernas titlar. Observera att vi har filtrerat bort funktionella ord (som 'the', 'a', 'this', etc.) samt 'COVID-19' och 'SARS-CoV-2' eftersom alla titlar innehöll dessa ord. Alla ordmoln uppdateras veckovis.
-
-**Källkod som används för att skapa ordmoln:** [Källkod som används för att skapa visualiseringarna](https://github.com/ScilifelabDataCentre/pathogens-portal-visualisations/tree/main/Wordcloud).
-
-#### Alla publikationer
-
-
Vetenskapsrådet:

SciLifeLab/KAW:

Horizon 2020:


Vetenskapsrådet:

SciLifeLab/KAW:

Horizon 2020:

{{ .Name }}
if (idValue === 'covid_publications') { matchingEbiEntry = findMatchingEntry(ebiEntries,'source_page', 'publications'); } - // Special case for 'wastewater' dashboard card - else if (idValue === 'wastewater') { - matchingEbiEntry = findMostRecentEntry(ebiEntries,'source_page', 'wastewater'); - } // Regular case where idValue is included in the source page path else { matchingEbiEntry = findMatchingEntry(ebiEntries,'source_page', idValue); @@ -197,7 +193,7 @@{{ .Name }}
* Find a matching EBI entry based on the specified field and its value */ function findMatchingEntry(ebiEntries, fieldName, fieldValue) { - return ebiEntries.find(entry => entry.fields.find(field => field.name === fieldName && field.value.includes(fieldValue))); + return ebiEntries.find(entry => entry.fields.find(field => field.name === fieldName && field.value.replace(/\/$/, "").endsWith(fieldValue))); } /** diff --git a/layouts/partials/navbar.html b/layouts/partials/navbar.html index d5180f3b5..9e5fad935 100644 --- a/layouts/partials/navbar.html +++ b/layouts/partials/navbar.html @@ -208,6 +208,12 @@ Provsamlingsregistret{{ end }} @@ -324,6 +330,12 @@ Provsamlingsregistret