This tutorial shows how to use DeepTree algorithm to select feature genes from any gene list (e.g. all genes, or highly variable genes)
These scripts were developed for He et al. 2020 Nature
Highly variable genes are not just about genes supporting cell types and cell states, they also contain sporadic genes that contribute to the noise of the transcriptome. By using co-expression information extracted from hierarchical clustering, we are able to remove some of these noises to make the modular gene expression structure clearer in downstream analyses.
If you de-noise the gene list per each batch and then merge the gene list for combined analysis, your batch effect will also be mitigated. Mitigating batch effect may save weak cell-type signatures compared to hard batch-effect correction methods that may mask some useful features.
This tutorial is only for MATLAB users. The python version is highly recommended because it is compatible with Scanpy. Check out here
Download the whole folder, and then point MATLAB to this folder as one of customer scripts. This is the code for Mac Users:
addpath(genpath('/Users/path_to_your_downloaded_and_extracted_codes'));
PC and Linux users need to pay attention to the path especially forward slash versus backward slash
We will use Seurat package's sample data here with Matlab 2018a on Mac.
These commands are for Unix terminal
Extracting the file on Unix terminal (or double click on MatLab Finder)
tar -xzf pbmc3k_filtered_gene_bc_matrices.tar.gz
The data will be in a folder called "filtered_gene_bc_matrices/hg19". Go to that folder.
Below are commands for Matlab. Please make sure your current directory is correct (see picture below)

Then import matrixmarket data to DataMatrix, a type similar to CellDataSet used by Monocle and SeuratObject in Seurat, on R platform.
PMBCdm=read10XCount('matrix.mtx','genes.tsv','barcodes.tsv');
or, by default if you used CellRanger to generate matrix.mtx, genes.tsv and barcodes.tsv in the current folder
PMBCdm=read10XCount();
Log normalize the data
PMBC_perc=log2(PMBCdm./sum(PMBCdm)*10000+1);
Check Data Quality
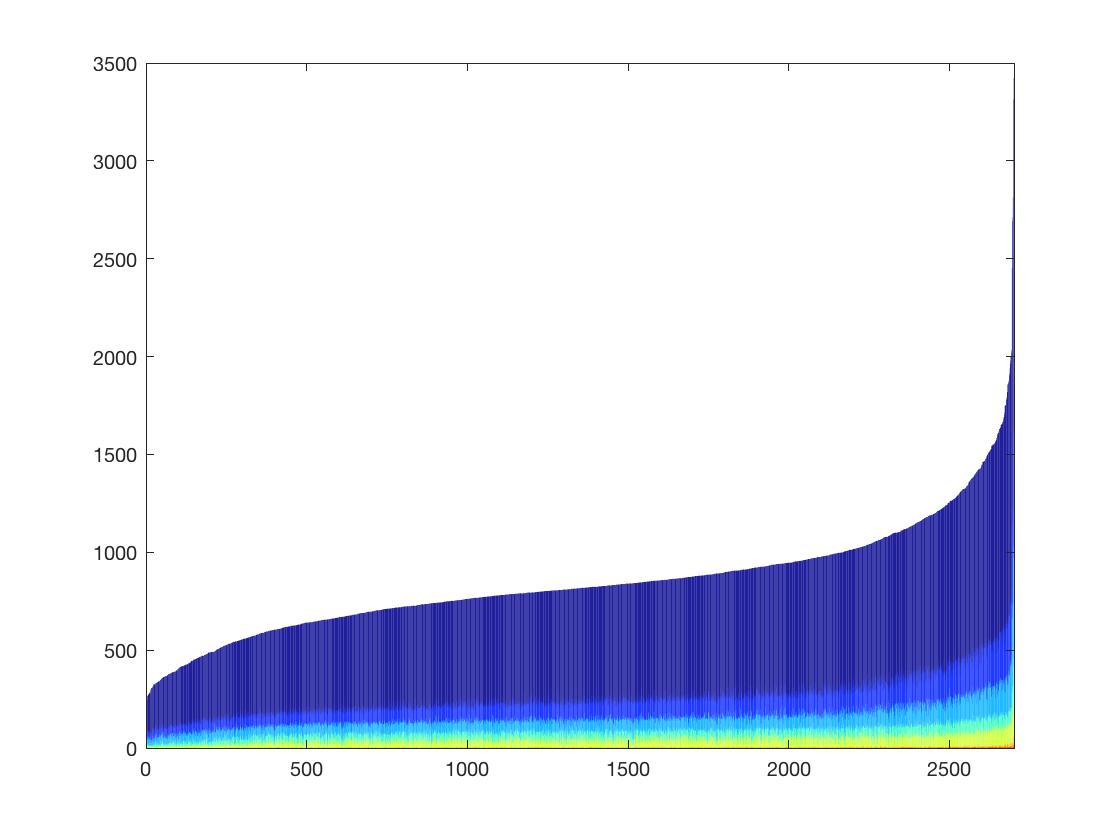
test=Gene10XCount(PMBCdm,1);
The plot shows the numbers of genes with 1 count/2 count/3count etc. The height of each bar represents the total number of genes detected in each cell.
Filter out low-quality cells (less than 500 genes detected) and low-quality genes (detected in less than 0.5% cells)
PMBC_perc_filtered=PMBC_perc(sum((PMBC_perc>0),2)>sum(sum(test)>500,2)/200,sum(test)>500);
Calculate high-dispersion genes
Dispersion=var(PMBC_perc_filtered,0,2)./mean(PMBC_perc_filtered,2);
[P I]=sort(Dispersion,'descend');
For the downstream analyses, following the codes shown above, a straightforward and naive method is provided here, which directly clusters the cells and comments on gene signature and dimensionality without bioinformatic tricks. More sophisticated methods are given below (please continue reading).
*If you want to remove mitochondria genes, you could use strnotcontain('_mt',XXXX) to remove all the genes whose names start with "mt".
Mainstream methods like Seurat calculates "highly variable genes" based on dispersion (variance/mean of log-transformed abundance estimate), which is only able to enrich cell type-specific genes and deplete cell-cycle associated and house-keeping genes, but still include a large number of sporadic noises. One key difference between real cell markers and noise is that real markers usually have their followers due to tight regulation of the gene network. So they sit in "modules" of co-regulated genes that are highly correlated with each other. This type of strong co-linearity can be captured as "deepest branches" of a phylogenetic tree of genes. Below I'm showing you how this Deep-tree analysis (python version can be found here) identifies markers. By doing this, you don't need to perform a PCA for cell clustering.
Here I use the top 4000 high-dispersion genes as an initial input.
PMBCDeepTree=DeepTreeCluster(PMBC_perc_filtered(I(1:4000),:),0.8,2)
This image shows a two-way clustergram of all the 4000 genes, most of which are sporadic noises without coherent patterns, seen as green areas with random red dots. The co-expressed genes, however, form deep branches in the tree, highlighted with distinguishable colors in the dendrogram. These 149 genes were extracted and used for a second clustering shown below.
which can be annotated interactively following Matlab Clustergram Manual to make it look like the following
In addition to the 8 clusters Seurat identified, small clusters (of as few as 3 cells) can also be identified, summing up to 11 cell types. In fact, high-dispersion cut favors identification of rare cell types but certain clustering methods may ignore small clusters of cells. Here we see that even for small clusters they may still be well supported by multiple marker genes that are co-expressed.
Promisingly, these 149 marker genes contain 8 of the 9 cherry-picked marker genes in Seurat Tutorial, confirming that real markers should be only a fraction of high-dispersion genes. CD3E is a marker for the most dominant cell type that was not captured by the "deep-tree" method but can still be retrived by looking for significantly different genes between defined clusters.
Here I use the 9 markers shown in Seurat Tutorial as an example to plot a 2-D heatmap.
SeuratMarkers={'ENSG00000156738_MS4A1','ENSG00000115523_GNLY','ENSG00000198851_CD3E','ENSG00000170458_CD14','ENSG00000179639_FCER1A','ENSG00000203747_FCGR3A','ENSG00000090382_LYZ','ENSG00000163736_PPBP','ENSG00000153563_CD8A'};
HeatMap(PMBC_perc_filtered(SeuratMarkers,get(PMBCDeepTree,'ColumnLabels')),'Standardize',2,'DisplayRange',2.5,'Colormap',colormap(jet),'Symmetric',true)
A HeatMap matching the cell order of the deep-tree clustergram is then generated.

If you only know the gene name (e.g. HOPX) but not the gene ID, you can look it up using this:
get(PMBCdm(strcontain('HOPX',get(PMBCdm,'RowNames')),:),'RowNames')
It will show the full name
If you want to use the clusters defined by hierarchical clustering, let's first drop down our manually assigned cell clusters from clustering analysis by right clicking the branches and saving them as Cluster1, Cluster2... Based on the coloring in the example image, the clusters are: Cluster 1 - Purple Cluster 2 - Orange Cluster 3 - Blue Cluster 4 - Dark Green Cluster 5 - Dark Blue Cluster 6 - Pink Cluster 7 - Cyan Cluster 8 - Red Cluster 9 - Green Cluster 10 - Yellow
ClusterID=repmat([0],size(get(PMBCDeepTree,'ColumnLabels')));
ClusterID(GetOrder(get(Cluster1,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=1;
ClusterID(GetOrder(get(Cluster2,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=2;
ClusterID(GetOrder(get(Cluster3,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=3;
ClusterID(GetOrder(get(Cluster4,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=4;
ClusterID(GetOrder(get(Cluster5,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=5;
ClusterID(GetOrder(get(Cluster6,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=6;
ClusterID(GetOrder(get(Cluster7,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=7;
ClusterID(GetOrder(get(Cluster8,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=8;
ClusterID(GetOrder(get(Cluster9,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=9;
ClusterID(GetOrder(get(Cluster10,'ColumnLabels'),get(PMBCDeepTree,'ColumnLabels')))=10;
Now calculate t-SNE coordinates and plot
mappedX=tsne(double(PMBC_perc_filtered(get(PMBCDeepTree,'RowLabels'),get(PMBCDeepTree,'ColumnLabels')))','Algorithm','barneshut','NumPCAComponents',0,'Distance','spearman','Perplexity',30);
You should right click mappedX in Workspace window to save it to a file. So that next time you can use mappedX=importdata(); to import it
figure
scatter(mappedX(:,1),mappedX(:,2),50,[0.8 0.8 0.8],'.')
 The t-SNE map may vary from run to run due to different seeds used.
But you should see that most cells fall in large "islands" while a few outliers are far apart.
The t-SNE map may vary from run to run due to different seeds used.
But you should see that most cells fall in large "islands" while a few outliers are far apart.
Now paint the t-SNE plot with cluster colors.
hold on
scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0.49 0.18 0.56],'.')
scatter(mappedX(ClusterID==2,1),mappedX(ClusterID==2,2),50,[0.85 0.33 0.1],'.')
scatter(mappedX(ClusterID==3,1),mappedX(ClusterID==3,2),50,[0 0.45 0.74],'.')
scatter(mappedX(ClusterID==4,1),mappedX(ClusterID==4,2),50,[0.47 0.67 0.19],'.')
scatter(mappedX(ClusterID==5,1),mappedX(ClusterID==5,2),50,[0.3 0.75 0.93],'.')
scatter(mappedX(ClusterID==6,1),mappedX(ClusterID==6,2),50,[1 0 1],'.')
scatter(mappedX(ClusterID==7,1),mappedX(ClusterID==7,2),50,[0 1 1],'.')
scatter(mappedX(ClusterID==8,1),mappedX(ClusterID==8,2),50,[1 0 0],'.')
scatter(mappedX(ClusterID==9,1),mappedX(ClusterID==9,2),50,[0 1 0],'.')
scatter(mappedX(ClusterID==10,1),mappedX(ClusterID==10,2),50,[1 1 0],'.')
Of course, you can do an unsupervised clustering using the top 10 PCs I mentioned above.
kmeans_colors=distinguishable_colors(10);
figure;scatter(mappedX(:,1),mappedX(:,2),50,kmeans_colors(kmeans(double(PMBC_perc_filtered(get(PMBCDeepTree,'RowLabels'),get(PMBCDeepTree,'ColumnLabels')))',9),:),'.');

figure
scatter(mappedX(:,1),mappedX(:,2),50,PMBC_perc_filtered('ENSG00000105369_CD79A',get(PMBCDeepTree,'ColumnLabels')),'.')
colormap(hot)
To export data matrix from Scanpy object, use this
adata.to_df().to_csv('matrix.csv')
Then import this matrix to MATLAB
PBMC=readtable('matrix.csv');
Then convert to DataMatrix format
PBMCdm=Table2DataMatrix(PBMC);
Then you can perform the normal workflow on MATLAB. Skip the normalization step if the matrix is already normalized.