diff --git a/README.md b/README.md
index 92a7400..81b1af6 100644
--- a/README.md
+++ b/README.md
@@ -52,7 +52,7 @@ Dispersion=var(PMBC_perc_filtered,0,2)./mean(PMBC_perc_filtered,2);
[P I]=sort(Dispersion,'descend');
```
-### Supervised Clustering and visualization
+### Supervised manual hierarchical clustering and visualization
PMBCClg=clustergram(PMBC_perc_filtered(I(1:2000),:),'Standardize',0,'DisplayRange',7.5,'Colormap',colormap(jet),'RowPDist','correlation','ColumnPDist','spearman','Symmetric',false,'linkage','complete', 'OptimalLeafOrder',false)
@@ -75,11 +75,12 @@ And then import this list to an array
```
GenesOfInterest=table2array(readtable('test.dat','ReadVariableNames',false,'Delimiter','\t'))
```
-Then plot into a HeatMap matching the cell order of your clustergram
+Then plot a HeatMap matching the cell order of your clustergram
```
HeatMap(PMBC_perc(GenesOfInterest,get(PMBCClg,'ColumnLabels')),'Standardize',2,'Colormap',colormap(jet),'Symmetric',true,'DisplayRange',2.5)
```
- +
+ +
*If you only know the gene name (e.g. HOPX) but not the gene ID, you can look it up using this:*
```
@@ -93,32 +94,37 @@ get(PMBCdm(strcontain('HOPX',get(PMBCdm,'RowNames')),:),'RowNames')
Before we do this, let's first drop down our manually assigned cell clusters from clustering analysis.
Based on the coloring in the example image, the clusters are:
```
-ClusterID=[repmat([1],1,451) repmat([0],1,44) repmat([2],1,137) repmat([3],1,23) repmat([0],1,664) repmat([4],1,119) repmat([0],1,20) repmat([5],1,313) repmat([0],1,195) repmat([6],1,54) repmat([7],1,202) repmat([8],1,244) repmat([0],1,15)];
+ClusterID=repmat([0],size(get(PMBCClg,'ColumnLabels')));
+ClusterID(GetOrder(get(Cluster1,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=1;
+ClusterID(GetOrder(get(Cluster2,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=2;
+ClusterID(GetOrder(get(Cluster3,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=3;
+ClusterID(GetOrder(get(Cluster4,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=4;
+ClusterID(GetOrder(get(Cluster5,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=5;
+ClusterID(GetOrder(get(Cluster6,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=6;
+ClusterID(GetOrder(get(Cluster7,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=7;
+ClusterID(GetOrder(get(Cluster8,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=8;
```
-Now inspect top 30 PCs for the normalized dataset (you can use absolute values, if you remove the function zscore())
+Now inspect top 30 PCs for the normalized dataset (you can use normalized values, if you use the function [zscore()](https://www.mathworks.com/help/stats/zscore.html?searchHighlight=zscore&s_tid=doc_srchtitle)).
```
-PCAPlot(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))')',get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:500),:),'RowNames'),ClusterID,100,30);
+[COEFF,SCORE,latent]=PCAPlot(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels'))),get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:2000),:),'RowNames'),ClusterID',100,30);
```
Among the outputs you get an image of PC scores, where each row represents each PC's scores across individual cells, ordered in the same way as the clustergram you generated.
-
+
*If you only know the gene name (e.g. HOPX) but not the gene ID, you can look it up using this:*
```
@@ -93,32 +94,37 @@ get(PMBCdm(strcontain('HOPX',get(PMBCdm,'RowNames')),:),'RowNames')
Before we do this, let's first drop down our manually assigned cell clusters from clustering analysis.
Based on the coloring in the example image, the clusters are:
```
-ClusterID=[repmat([1],1,451) repmat([0],1,44) repmat([2],1,137) repmat([3],1,23) repmat([0],1,664) repmat([4],1,119) repmat([0],1,20) repmat([5],1,313) repmat([0],1,195) repmat([6],1,54) repmat([7],1,202) repmat([8],1,244) repmat([0],1,15)];
+ClusterID=repmat([0],size(get(PMBCClg,'ColumnLabels')));
+ClusterID(GetOrder(get(Cluster1,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=1;
+ClusterID(GetOrder(get(Cluster2,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=2;
+ClusterID(GetOrder(get(Cluster3,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=3;
+ClusterID(GetOrder(get(Cluster4,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=4;
+ClusterID(GetOrder(get(Cluster5,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=5;
+ClusterID(GetOrder(get(Cluster6,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=6;
+ClusterID(GetOrder(get(Cluster7,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=7;
+ClusterID(GetOrder(get(Cluster8,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=8;
```
-Now inspect top 30 PCs for the normalized dataset (you can use absolute values, if you remove the function zscore())
+Now inspect top 30 PCs for the normalized dataset (you can use normalized values, if you use the function [zscore()](https://www.mathworks.com/help/stats/zscore.html?searchHighlight=zscore&s_tid=doc_srchtitle)).
```
-PCAPlot(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))')',get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:500),:),'RowNames'),ClusterID,100,30);
+[COEFF,SCORE,latent]=PCAPlot(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels'))),get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:2000),:),'RowNames'),ClusterID',100,30);
```
Among the outputs you get an image of PC scores, where each row represents each PC's scores across individual cells, ordered in the same way as the clustergram you generated.
- +
+ -Based on this diagram, we see that only the top 9 PCs show cell-type specificity. So we only use these 9 PCs to plot t-SNE.
+Based on this diagram, we see that only the top 10 PCs show cell-type specificity. So we only use these 10 PCs to plot t-SNE.
Now calculate t-SNE coordinates and plot
```
-mappedX=tsne(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))'),'Algorithm','barneshut','NumPCAComponents',9,'Distance','spearman','Perplexity',30);
+mappedX=tsne(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels')))','Algorithm','barneshut','NumPCAComponents',10,'Distance','spearman','Perplexity',30);
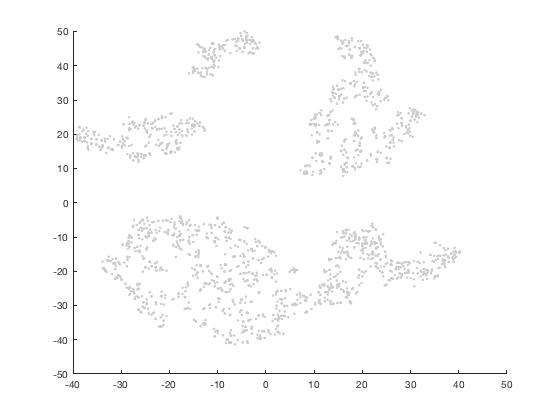
scatter(mappedX(:,1),mappedX(:,2),50,[0.8 0.8 0.8],'.')
```
-
+
*You should right click mappedX in Workspace window to save it to a file. So that next time you can use mappedX=importdata(); to import it*
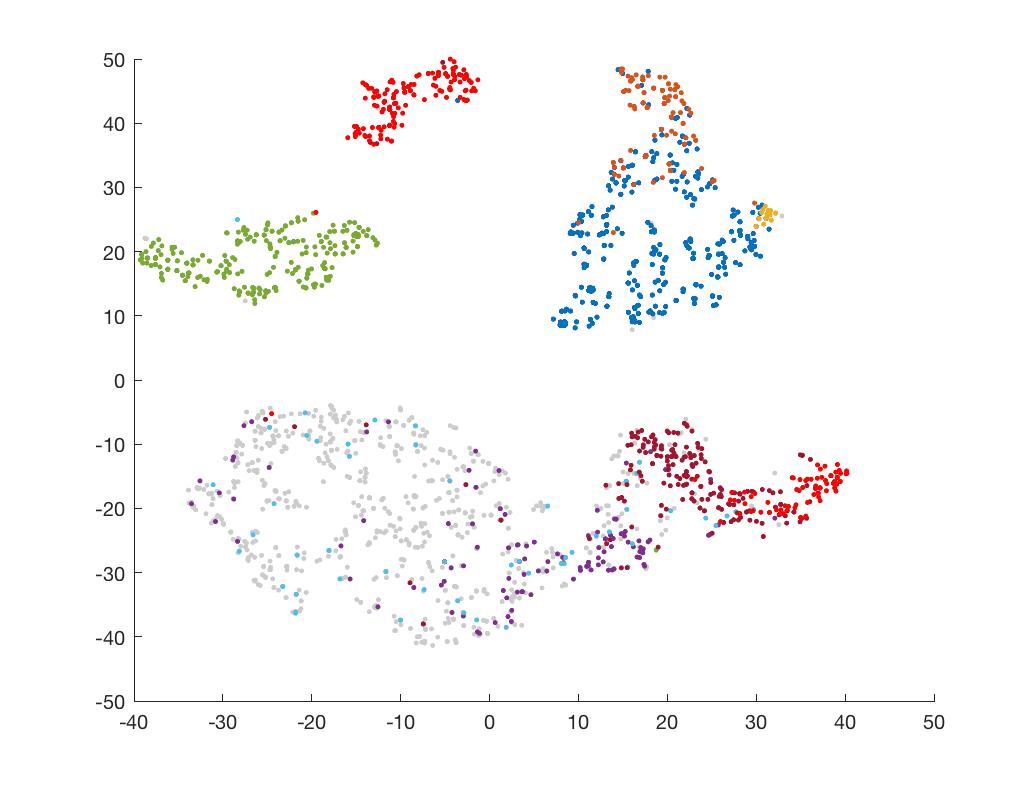
Now paint the t-SNE plot with cluster colors.
```
hold on
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0 1],'.')
scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0.45 0.74],'.')
scatter(mappedX(ClusterID==2,1),mappedX(ClusterID==2,2),50,[0.85 0.33 0.1],'.')
scatter(mappedX(ClusterID==3,1),mappedX(ClusterID==3,2),50,[0.93 0.69 0.13],'.')
@@ -128,7 +134,15 @@ scatter(mappedX(ClusterID==6,1),mappedX(ClusterID==6,2),50,[0.3 0.75 0.93],'.')
scatter(mappedX(ClusterID==7,1),mappedX(ClusterID==7,2),50,[0.64 0.08 0.18],'.')
scatter(mappedX(ClusterID==8,1),mappedX(ClusterID==8,2),50,[1 0 0],'.')
```
-
+
+
+
+#### Unsupervised k-means clustering
+Of course, you can do an unsupervised clustering using the top 10 PCs I mentioned above.
+kmeans_colors=distinguishable_colors(10);
+scatter(mappedX(:,1),mappedX(:,2),50,kmeans_colors(kmeans(SCORE(:,1:10),10),:),'.');
+
+
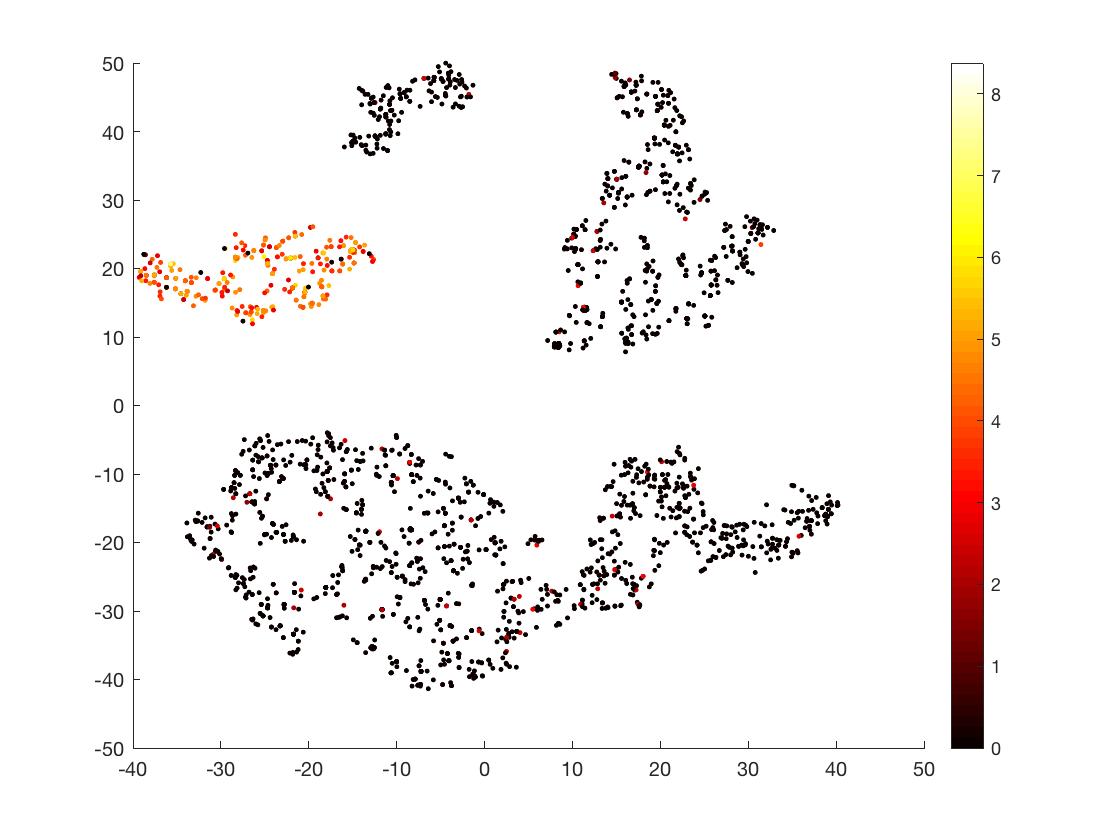
#### Check expression of a specific gene
```
@@ -136,7 +150,7 @@ figure
scatter(mappedX(:,1),mappedX(:,2),50,PMBC_perc('ENSG00000105369_CD79A',get(PMBCClg,'ColumnLabels')),'.')
colormap(hot)
```
-
+
## Discussions
### Parameter optimizations
-Based on this diagram, we see that only the top 9 PCs show cell-type specificity. So we only use these 9 PCs to plot t-SNE.
+Based on this diagram, we see that only the top 10 PCs show cell-type specificity. So we only use these 10 PCs to plot t-SNE.
Now calculate t-SNE coordinates and plot
```
-mappedX=tsne(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))'),'Algorithm','barneshut','NumPCAComponents',9,'Distance','spearman','Perplexity',30);
+mappedX=tsne(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels')))','Algorithm','barneshut','NumPCAComponents',10,'Distance','spearman','Perplexity',30);
scatter(mappedX(:,1),mappedX(:,2),50,[0.8 0.8 0.8],'.')
```
-
+
*You should right click mappedX in Workspace window to save it to a file. So that next time you can use mappedX=importdata(); to import it*
Now paint the t-SNE plot with cluster colors.
```
hold on
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0 1],'.')
scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0.45 0.74],'.')
scatter(mappedX(ClusterID==2,1),mappedX(ClusterID==2,2),50,[0.85 0.33 0.1],'.')
scatter(mappedX(ClusterID==3,1),mappedX(ClusterID==3,2),50,[0.93 0.69 0.13],'.')
@@ -128,7 +134,15 @@ scatter(mappedX(ClusterID==6,1),mappedX(ClusterID==6,2),50,[0.3 0.75 0.93],'.')
scatter(mappedX(ClusterID==7,1),mappedX(ClusterID==7,2),50,[0.64 0.08 0.18],'.')
scatter(mappedX(ClusterID==8,1),mappedX(ClusterID==8,2),50,[1 0 0],'.')
```
-
+
+
+
+#### Unsupervised k-means clustering
+Of course, you can do an unsupervised clustering using the top 10 PCs I mentioned above.
+kmeans_colors=distinguishable_colors(10);
+scatter(mappedX(:,1),mappedX(:,2),50,kmeans_colors(kmeans(SCORE(:,1:10),10),:),'.');
+
+
#### Check expression of a specific gene
```
@@ -136,7 +150,7 @@ figure
scatter(mappedX(:,1),mappedX(:,2),50,PMBC_perc('ENSG00000105369_CD79A',get(PMBCClg,'ColumnLabels')),'.')
colormap(hot)
```
-
+
## Discussions
### Parameter optimizations
 +
+ +
*If you only know the gene name (e.g. HOPX) but not the gene ID, you can look it up using this:*
```
@@ -93,32 +94,37 @@ get(PMBCdm(strcontain('HOPX',get(PMBCdm,'RowNames')),:),'RowNames')
Before we do this, let's first drop down our manually assigned cell clusters from clustering analysis.
Based on the coloring in the example image, the clusters are:
```
-ClusterID=[repmat([1],1,451) repmat([0],1,44) repmat([2],1,137) repmat([3],1,23) repmat([0],1,664) repmat([4],1,119) repmat([0],1,20) repmat([5],1,313) repmat([0],1,195) repmat([6],1,54) repmat([7],1,202) repmat([8],1,244) repmat([0],1,15)];
+ClusterID=repmat([0],size(get(PMBCClg,'ColumnLabels')));
+ClusterID(GetOrder(get(Cluster1,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=1;
+ClusterID(GetOrder(get(Cluster2,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=2;
+ClusterID(GetOrder(get(Cluster3,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=3;
+ClusterID(GetOrder(get(Cluster4,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=4;
+ClusterID(GetOrder(get(Cluster5,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=5;
+ClusterID(GetOrder(get(Cluster6,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=6;
+ClusterID(GetOrder(get(Cluster7,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=7;
+ClusterID(GetOrder(get(Cluster8,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=8;
```
-Now inspect top 30 PCs for the normalized dataset (you can use absolute values, if you remove the function zscore())
+Now inspect top 30 PCs for the normalized dataset (you can use normalized values, if you use the function [zscore()](https://www.mathworks.com/help/stats/zscore.html?searchHighlight=zscore&s_tid=doc_srchtitle)).
```
-PCAPlot(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))')',get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:500),:),'RowNames'),ClusterID,100,30);
+[COEFF,SCORE,latent]=PCAPlot(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels'))),get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:2000),:),'RowNames'),ClusterID',100,30);
```
Among the outputs you get an image of PC scores, where each row represents each PC's scores across individual cells, ordered in the same way as the clustergram you generated.
-
+
*If you only know the gene name (e.g. HOPX) but not the gene ID, you can look it up using this:*
```
@@ -93,32 +94,37 @@ get(PMBCdm(strcontain('HOPX',get(PMBCdm,'RowNames')),:),'RowNames')
Before we do this, let's first drop down our manually assigned cell clusters from clustering analysis.
Based on the coloring in the example image, the clusters are:
```
-ClusterID=[repmat([1],1,451) repmat([0],1,44) repmat([2],1,137) repmat([3],1,23) repmat([0],1,664) repmat([4],1,119) repmat([0],1,20) repmat([5],1,313) repmat([0],1,195) repmat([6],1,54) repmat([7],1,202) repmat([8],1,244) repmat([0],1,15)];
+ClusterID=repmat([0],size(get(PMBCClg,'ColumnLabels')));
+ClusterID(GetOrder(get(Cluster1,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=1;
+ClusterID(GetOrder(get(Cluster2,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=2;
+ClusterID(GetOrder(get(Cluster3,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=3;
+ClusterID(GetOrder(get(Cluster4,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=4;
+ClusterID(GetOrder(get(Cluster5,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=5;
+ClusterID(GetOrder(get(Cluster6,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=6;
+ClusterID(GetOrder(get(Cluster7,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=7;
+ClusterID(GetOrder(get(Cluster8,'ColumnLabels'),get(PMBCClg,'ColumnLabels')))=8;
```
-Now inspect top 30 PCs for the normalized dataset (you can use absolute values, if you remove the function zscore())
+Now inspect top 30 PCs for the normalized dataset (you can use normalized values, if you use the function [zscore()](https://www.mathworks.com/help/stats/zscore.html?searchHighlight=zscore&s_tid=doc_srchtitle)).
```
-PCAPlot(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))')',get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:500),:),'RowNames'),ClusterID,100,30);
+[COEFF,SCORE,latent]=PCAPlot(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels'))),get(PMBCClg,'ColumnLabels'),get(PMBC_perc_filtered(I(1:2000),:),'RowNames'),ClusterID',100,30);
```
Among the outputs you get an image of PC scores, where each row represents each PC's scores across individual cells, ordered in the same way as the clustergram you generated.
- +
+ -Based on this diagram, we see that only the top 9 PCs show cell-type specificity. So we only use these 9 PCs to plot t-SNE.
+Based on this diagram, we see that only the top 10 PCs show cell-type specificity. So we only use these 10 PCs to plot t-SNE.
Now calculate t-SNE coordinates and plot
```
-mappedX=tsne(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))'),'Algorithm','barneshut','NumPCAComponents',9,'Distance','spearman','Perplexity',30);
+mappedX=tsne(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels')))','Algorithm','barneshut','NumPCAComponents',10,'Distance','spearman','Perplexity',30);
scatter(mappedX(:,1),mappedX(:,2),50,[0.8 0.8 0.8],'.')
```
-
+
*You should right click mappedX in Workspace window to save it to a file. So that next time you can use mappedX=importdata(); to import it*
Now paint the t-SNE plot with cluster colors.
```
hold on
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0 1],'.')
scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0.45 0.74],'.')
scatter(mappedX(ClusterID==2,1),mappedX(ClusterID==2,2),50,[0.85 0.33 0.1],'.')
scatter(mappedX(ClusterID==3,1),mappedX(ClusterID==3,2),50,[0.93 0.69 0.13],'.')
@@ -128,7 +134,15 @@ scatter(mappedX(ClusterID==6,1),mappedX(ClusterID==6,2),50,[0.3 0.75 0.93],'.')
scatter(mappedX(ClusterID==7,1),mappedX(ClusterID==7,2),50,[0.64 0.08 0.18],'.')
scatter(mappedX(ClusterID==8,1),mappedX(ClusterID==8,2),50,[1 0 0],'.')
```
-
+
+
+
+#### Unsupervised k-means clustering
+Of course, you can do an unsupervised clustering using the top 10 PCs I mentioned above.
+kmeans_colors=distinguishable_colors(10);
+scatter(mappedX(:,1),mappedX(:,2),50,kmeans_colors(kmeans(SCORE(:,1:10),10),:),'.');
+
+
#### Check expression of a specific gene
```
@@ -136,7 +150,7 @@ figure
scatter(mappedX(:,1),mappedX(:,2),50,PMBC_perc('ENSG00000105369_CD79A',get(PMBCClg,'ColumnLabels')),'.')
colormap(hot)
```
-
+
## Discussions
### Parameter optimizations
-Based on this diagram, we see that only the top 9 PCs show cell-type specificity. So we only use these 9 PCs to plot t-SNE.
+Based on this diagram, we see that only the top 10 PCs show cell-type specificity. So we only use these 10 PCs to plot t-SNE.
Now calculate t-SNE coordinates and plot
```
-mappedX=tsne(zscore(double(PMBC_perc_filtered(I(1:500),get(PMBCClg,'ColumnLabels')))'),'Algorithm','barneshut','NumPCAComponents',9,'Distance','spearman','Perplexity',30);
+mappedX=tsne(double(PMBC_perc_filtered(I(1:2000),get(PMBCClg,'ColumnLabels')))','Algorithm','barneshut','NumPCAComponents',10,'Distance','spearman','Perplexity',30);
scatter(mappedX(:,1),mappedX(:,2),50,[0.8 0.8 0.8],'.')
```
-
+
*You should right click mappedX in Workspace window to save it to a file. So that next time you can use mappedX=importdata(); to import it*
Now paint the t-SNE plot with cluster colors.
```
hold on
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),10,[0 0 1],'.')
-scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0 1],'.')
scatter(mappedX(ClusterID==1,1),mappedX(ClusterID==1,2),50,[0 0.45 0.74],'.')
scatter(mappedX(ClusterID==2,1),mappedX(ClusterID==2,2),50,[0.85 0.33 0.1],'.')
scatter(mappedX(ClusterID==3,1),mappedX(ClusterID==3,2),50,[0.93 0.69 0.13],'.')
@@ -128,7 +134,15 @@ scatter(mappedX(ClusterID==6,1),mappedX(ClusterID==6,2),50,[0.3 0.75 0.93],'.')
scatter(mappedX(ClusterID==7,1),mappedX(ClusterID==7,2),50,[0.64 0.08 0.18],'.')
scatter(mappedX(ClusterID==8,1),mappedX(ClusterID==8,2),50,[1 0 0],'.')
```
-
+
+
+
+#### Unsupervised k-means clustering
+Of course, you can do an unsupervised clustering using the top 10 PCs I mentioned above.
+kmeans_colors=distinguishable_colors(10);
+scatter(mappedX(:,1),mappedX(:,2),50,kmeans_colors(kmeans(SCORE(:,1:10),10),:),'.');
+
+
#### Check expression of a specific gene
```
@@ -136,7 +150,7 @@ figure
scatter(mappedX(:,1),mappedX(:,2),50,PMBC_perc('ENSG00000105369_CD79A',get(PMBCClg,'ColumnLabels')),'.')
colormap(hot)
```
-
+
## Discussions
### Parameter optimizations