Gregory Way, 2020
In the following module, we test the ability of Cell Painting to predict cell health readouts.
The 3.train analysis module trains all models to predict cell health readouts.
The module stores data, software, results, figures, interpretations, and computational infrastructure to reproduce the analysis.
Both the Cell Painting and Cell Health assays were acquired on the same cell lines (A549, ES2, and HCC44) in the same perturbations. The assays were acquired from a CRISPR experiment in which 59 genes were knocked out with, in most cases, 2 different CRISPR guides.
First, we aggregated data at the treatment level (aggregating replicate wells together) forming consensus profiles. This procedure results in matched cell health readouts (y) with Cell Painting profiles (X). We randomly split the data (n = 357) into 85% training and 15% testing sets.
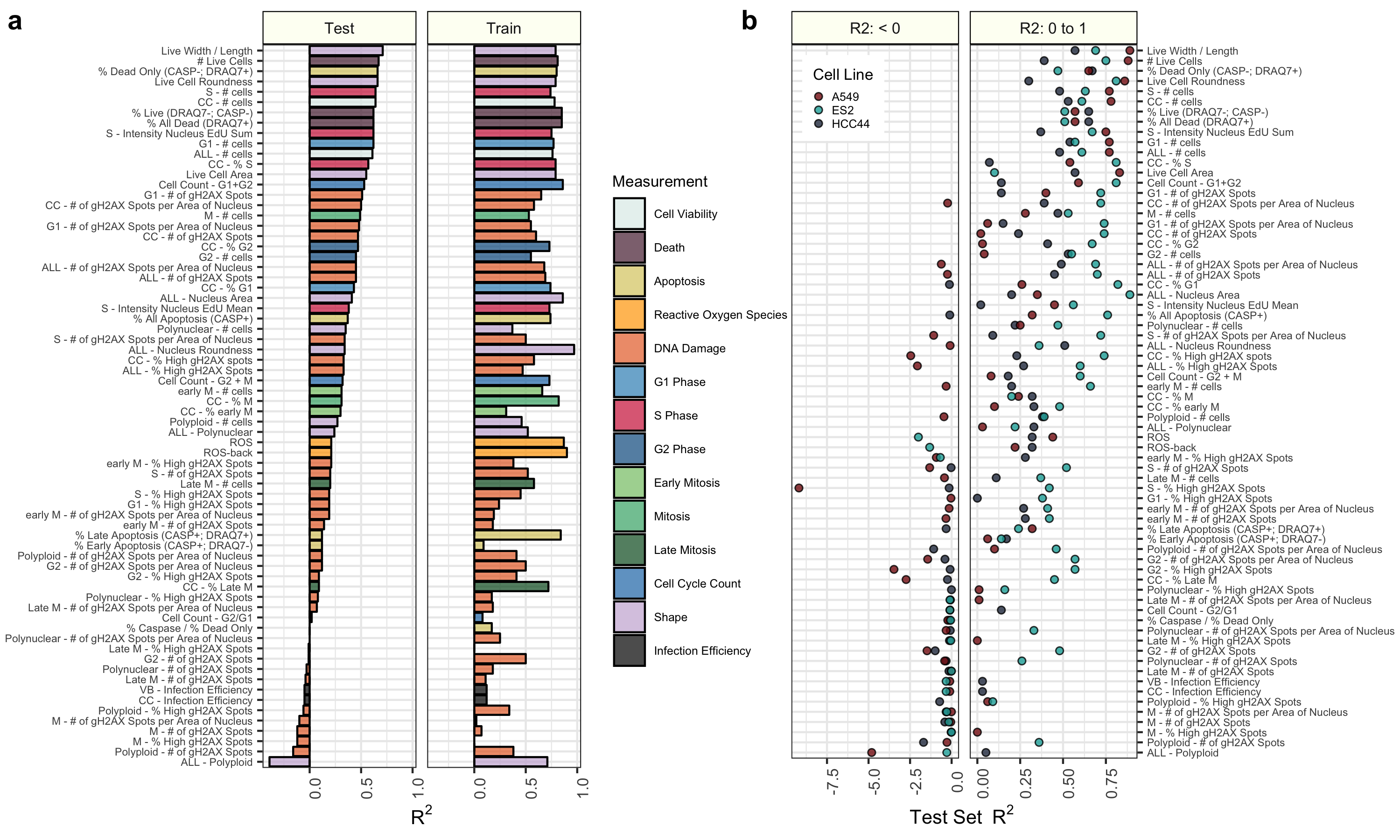
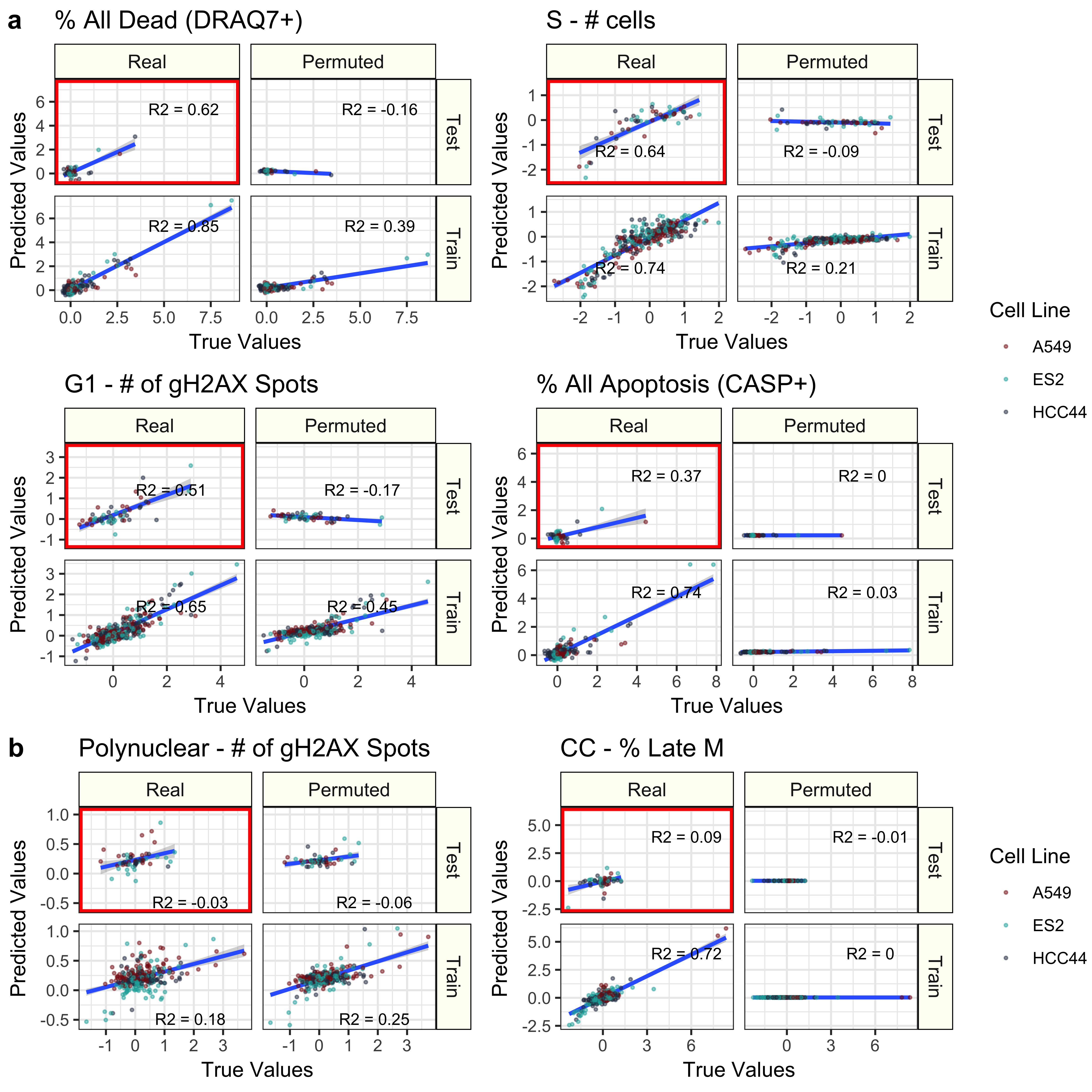
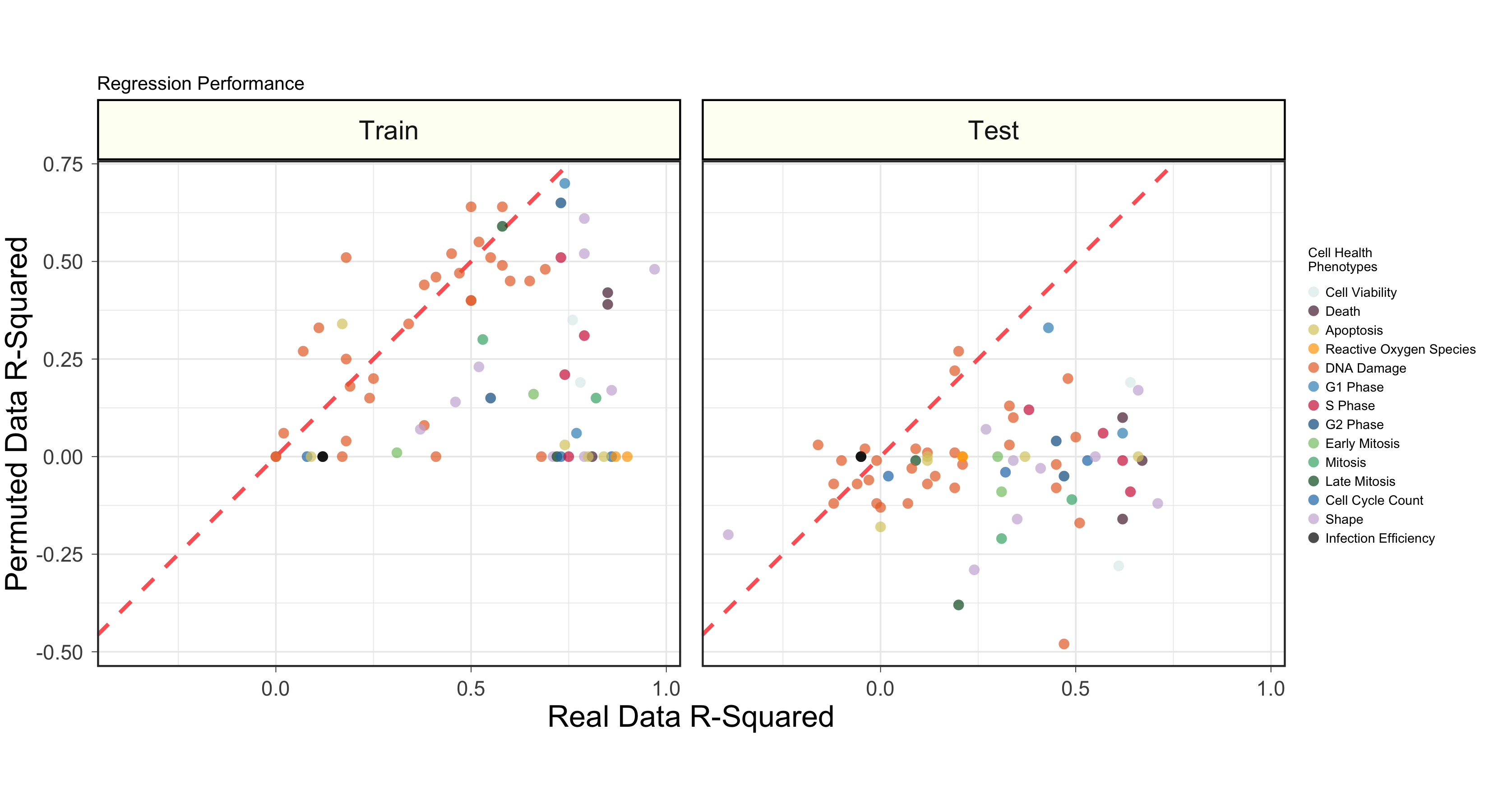
We then independently trained regression models to predict each of the 70 cell health readouts. We also trained models again using randomly permuted Cell Painting data.
Many cell health readouts can be predicted with high accuracy in a testing set.
Some cell health readouts can be predicted exceptionally well, but others cannot be captured with our current approach.
Cell painting can be used to predict cell health readouts better than models trained with random.
More specific results exploring performance for all models and all cell health variables are presented in the figures/ folder.