diff --git a/DESCRIPTION b/DESCRIPTION
index 9c6378ca..d0b3c980 100644
--- a/DESCRIPTION
+++ b/DESCRIPTION
@@ -1,7 +1,7 @@
Package: flextable
Type: Package
Title: Functions for Tabular Reporting
-Version: 0.9.5.003
+Version: 0.9.5.004
Authors@R: c(
person("David", "Gohel", role = c("aut", "cre"),
email = "david.gohel@ardata.fr"),
diff --git a/R/augment_rows.R b/R/augment_rows.R
index 2a511400..c0694a4c 100644
--- a/R/augment_rows.R
+++ b/R/augment_rows.R
@@ -1008,29 +1008,24 @@ set_footer_df <- function(x, mapping = NULL, key = "col_keys") {
#' @importFrom data.table tstrsplit
#' @export
-#' @title Separate collapsed colnames into multiple rows
-#' @description If your variable names contain
-#' multiple delimited labels, they will be separated
-#' and placed in their own rows.
+#' @title Split column names using a separator into multiple rows
+#' @description This function is used to separate and place individual
+#' labels in their own rows if your variable names contain multiple
+#' delimited labels.
#' \if{html}{\out{
#'  #' }}
#' @param x a flextable object
-#' @param opts optional treatments to apply
-#' to the resulting header part as a character
-#' vector with multiple supported values.
-#'
-#' The supported values are:
-#'
-#' * "span-top": span empty cells with the
-#' first non empty cell, this operation is made
-#' column by column.
-#' * "center-hspan": center the cells that are
-#' horizontally spanned.
-#' * "bottom-vspan": bottom align the cells treated
-#' when "span-top" is applied.
-#' * "default-theme": apply to the new header part
-#' the theme set in `set_flextable_defaults(theme_fun = ...)`.
+#' @param opts Optional treatments to apply to the resulting header part.
+#' This should be a character vector with support for multiple values.
+#'
+#' Supported values include:
+#'
+#' - "span-top": This operation spans empty cells with the first non-empty
+#' cell, applied column by column.
+#' - "center-hspan": Center the cells that are horizontally spanned.
+#' - "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+#' - "default-theme": Applies the theme set in `set_flextable_defaults(theme_fun = ...)` to the new header part.
#' @param split a regular expression (unless `fixed = TRUE`)
#' to use for splitting.
#' @param fixed logical. If TRUE match `split` exactly,
@@ -1105,11 +1100,12 @@ separate_header <- function(x,
for (j in seq_len(nrow(ref_list))) {
if (ref_list[j, 1]) {
to <- rle(ref_list[j, ])$lengths[1] + 1
-
- x <- merge_at(
- x = x, i = seq(1, to), j = j,
- part = "header"
- )
+ if (all(x$header$spans$rows[seq(1, to), j] %in% 1)) {#can be v-merged
+ x <- merge_at(
+ x = x, i = seq(1, to), j = j,
+ part = "header"
+ )
+ }
if ("bottom-vspan" %in% opts) {
x <- valign(
diff --git a/man/separate_header.Rd b/man/separate_header.Rd
index a318e409..cb4a86a1 100644
--- a/man/separate_header.Rd
+++ b/man/separate_header.Rd
@@ -2,7 +2,7 @@
% Please edit documentation in R/augment_rows.R
\name{separate_header}
\alias{separate_header}
-\title{Separate collapsed colnames into multiple rows}
+\title{Split column names using a separator into multiple rows}
\usage{
separate_header(
x,
@@ -14,21 +14,16 @@ separate_header(
\arguments{
\item{x}{a flextable object}
-\item{opts}{optional treatments to apply
-to the resulting header part as a character
-vector with multiple supported values.
+\item{opts}{Optional treatments to apply to the resulting header part.
+This should be a character vector with support for multiple values.
-The supported values are:
+Supported values include:
\itemize{
-\item "span-top": span empty cells with the
-first non empty cell, this operation is made
-column by column.
-\item "center-hspan": center the cells that are
-horizontally spanned.
-\item "bottom-vspan": bottom align the cells treated
-when "span-top" is applied.
-\item "default-theme": apply to the new header part
-the theme set in \code{set_flextable_defaults(theme_fun = ...)}.
+\item "span-top": This operation spans empty cells with the first non-empty
+cell, applied column by column.
+\item "center-hspan": Center the cells that are horizontally spanned.
+\item "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+\item "default-theme": Applies the theme set in \code{set_flextable_defaults(theme_fun = ...)} to the new header part.
}}
\item{split}{a regular expression (unless \code{fixed = TRUE})
@@ -38,9 +33,9 @@ to use for splitting.}
otherwise use regular expressions.}
}
\description{
-If your variable names contain
-multiple delimited labels, they will be separated
-and placed in their own rows.
+This function is used to separate and place individual
+labels in their own rows if your variable names contain multiple
+delimited labels.
\if{html}{\out{
#' }}
#' @param x a flextable object
-#' @param opts optional treatments to apply
-#' to the resulting header part as a character
-#' vector with multiple supported values.
-#'
-#' The supported values are:
-#'
-#' * "span-top": span empty cells with the
-#' first non empty cell, this operation is made
-#' column by column.
-#' * "center-hspan": center the cells that are
-#' horizontally spanned.
-#' * "bottom-vspan": bottom align the cells treated
-#' when "span-top" is applied.
-#' * "default-theme": apply to the new header part
-#' the theme set in `set_flextable_defaults(theme_fun = ...)`.
+#' @param opts Optional treatments to apply to the resulting header part.
+#' This should be a character vector with support for multiple values.
+#'
+#' Supported values include:
+#'
+#' - "span-top": This operation spans empty cells with the first non-empty
+#' cell, applied column by column.
+#' - "center-hspan": Center the cells that are horizontally spanned.
+#' - "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+#' - "default-theme": Applies the theme set in `set_flextable_defaults(theme_fun = ...)` to the new header part.
#' @param split a regular expression (unless `fixed = TRUE`)
#' to use for splitting.
#' @param fixed logical. If TRUE match `split` exactly,
@@ -1105,11 +1100,12 @@ separate_header <- function(x,
for (j in seq_len(nrow(ref_list))) {
if (ref_list[j, 1]) {
to <- rle(ref_list[j, ])$lengths[1] + 1
-
- x <- merge_at(
- x = x, i = seq(1, to), j = j,
- part = "header"
- )
+ if (all(x$header$spans$rows[seq(1, to), j] %in% 1)) {#can be v-merged
+ x <- merge_at(
+ x = x, i = seq(1, to), j = j,
+ part = "header"
+ )
+ }
if ("bottom-vspan" %in% opts) {
x <- valign(
diff --git a/man/separate_header.Rd b/man/separate_header.Rd
index a318e409..cb4a86a1 100644
--- a/man/separate_header.Rd
+++ b/man/separate_header.Rd
@@ -2,7 +2,7 @@
% Please edit documentation in R/augment_rows.R
\name{separate_header}
\alias{separate_header}
-\title{Separate collapsed colnames into multiple rows}
+\title{Split column names using a separator into multiple rows}
\usage{
separate_header(
x,
@@ -14,21 +14,16 @@ separate_header(
\arguments{
\item{x}{a flextable object}
-\item{opts}{optional treatments to apply
-to the resulting header part as a character
-vector with multiple supported values.
+\item{opts}{Optional treatments to apply to the resulting header part.
+This should be a character vector with support for multiple values.
-The supported values are:
+Supported values include:
\itemize{
-\item "span-top": span empty cells with the
-first non empty cell, this operation is made
-column by column.
-\item "center-hspan": center the cells that are
-horizontally spanned.
-\item "bottom-vspan": bottom align the cells treated
-when "span-top" is applied.
-\item "default-theme": apply to the new header part
-the theme set in \code{set_flextable_defaults(theme_fun = ...)}.
+\item "span-top": This operation spans empty cells with the first non-empty
+cell, applied column by column.
+\item "center-hspan": Center the cells that are horizontally spanned.
+\item "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+\item "default-theme": Applies the theme set in \code{set_flextable_defaults(theme_fun = ...)} to the new header part.
}}
\item{split}{a regular expression (unless \code{fixed = TRUE})
@@ -38,9 +33,9 @@ to use for splitting.}
otherwise use regular expressions.}
}
\description{
-If your variable names contain
-multiple delimited labels, they will be separated
-and placed in their own rows.
+This function is used to separate and place individual
+labels in their own rows if your variable names contain multiple
+delimited labels.
\if{html}{\out{
 }}
diff --git a/tests/testthat/test-merge.R b/tests/testthat/test-merge.R
index 3b45390d..c9dc90d5 100644
--- a/tests/testthat/test-merge.R
+++ b/tests/testthat/test-merge.R
@@ -55,3 +55,34 @@ test_that("merged cells can be un-merged", {
ft <- merge_none(ft)
expect_true(all(ft$body$spans$columns == 1))
})
+
+test_that("separate_header", {
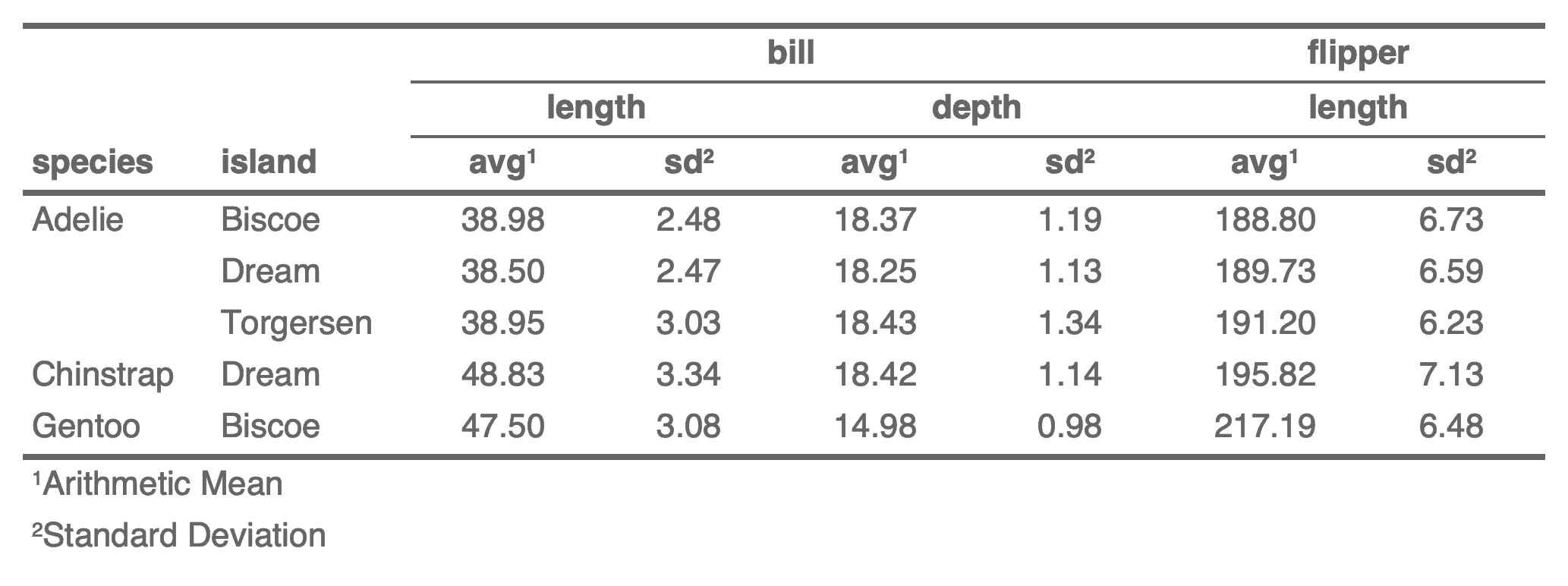
+ x <- data.frame(
+ Species = as.factor(c("setosa", "versicolor", "virginica")),
+ Sepal.Length_mean_zzz = c(5.006, 5.936, 6.588),
+ Sepal.Length_sd = c(0.35249, 0.51617, 0.63588),
+ Sepal.Width_mean = c(3.428, 2.77, 2.974),
+ Sepal.Width_sd_sfsf_dsfsdf = c(0.37906, 0.3138, 0.3225),
+ Petal.Length_mean = c(1.462, 4.26, 5.552),
+ Petal.Length_sd = c(0.17366, 0.46991, 0.55189),
+ Petal.Width_mean = c(0.246, 1.326, 2.026),
+ Petal.Width_sd = c(0.10539, 0.19775, 0.27465)
+ )
+
+ ft_1 <- flextable(x)
+ ft_1 <- separate_header(x = ft_1,
+ opts = c("span-top", "bottom-vspan")
+ )
+ header_txt <- flextable:::fortify_run(ft_1) |>
+ subset(.part %in% "header")
+ expect_equal(
+ object = header_txt$txt,
+ expected =
+ c("Species", "Sepal", "Sepal", "Sepal", "Sepal", "Petal", "Petal",
+ "Petal", "Petal", "", "Length", "Length", "Width", "Width", "Length",
+ "Length", "Width", "Width", "", "mean", "sd", "mean", "sd", "mean",
+ "sd", "mean", "sd", "", "zzz", "", "", "sfsf", "", "", "", "",
+ "", "", "", "", "dsfsdf", "", "", "", "")
+ )
+
+})
}}
diff --git a/tests/testthat/test-merge.R b/tests/testthat/test-merge.R
index 3b45390d..c9dc90d5 100644
--- a/tests/testthat/test-merge.R
+++ b/tests/testthat/test-merge.R
@@ -55,3 +55,34 @@ test_that("merged cells can be un-merged", {
ft <- merge_none(ft)
expect_true(all(ft$body$spans$columns == 1))
})
+
+test_that("separate_header", {
+ x <- data.frame(
+ Species = as.factor(c("setosa", "versicolor", "virginica")),
+ Sepal.Length_mean_zzz = c(5.006, 5.936, 6.588),
+ Sepal.Length_sd = c(0.35249, 0.51617, 0.63588),
+ Sepal.Width_mean = c(3.428, 2.77, 2.974),
+ Sepal.Width_sd_sfsf_dsfsdf = c(0.37906, 0.3138, 0.3225),
+ Petal.Length_mean = c(1.462, 4.26, 5.552),
+ Petal.Length_sd = c(0.17366, 0.46991, 0.55189),
+ Petal.Width_mean = c(0.246, 1.326, 2.026),
+ Petal.Width_sd = c(0.10539, 0.19775, 0.27465)
+ )
+
+ ft_1 <- flextable(x)
+ ft_1 <- separate_header(x = ft_1,
+ opts = c("span-top", "bottom-vspan")
+ )
+ header_txt <- flextable:::fortify_run(ft_1) |>
+ subset(.part %in% "header")
+ expect_equal(
+ object = header_txt$txt,
+ expected =
+ c("Species", "Sepal", "Sepal", "Sepal", "Sepal", "Petal", "Petal",
+ "Petal", "Petal", "", "Length", "Length", "Width", "Width", "Length",
+ "Length", "Width", "Width", "", "mean", "sd", "mean", "sd", "mean",
+ "sd", "mean", "sd", "", "zzz", "", "", "sfsf", "", "", "", "",
+ "", "", "", "", "dsfsdf", "", "", "", "")
+ )
+
+})
 #' }}
#' @param x a flextable object
-#' @param opts optional treatments to apply
-#' to the resulting header part as a character
-#' vector with multiple supported values.
-#'
-#' The supported values are:
-#'
-#' * "span-top": span empty cells with the
-#' first non empty cell, this operation is made
-#' column by column.
-#' * "center-hspan": center the cells that are
-#' horizontally spanned.
-#' * "bottom-vspan": bottom align the cells treated
-#' when "span-top" is applied.
-#' * "default-theme": apply to the new header part
-#' the theme set in `set_flextable_defaults(theme_fun = ...)`.
+#' @param opts Optional treatments to apply to the resulting header part.
+#' This should be a character vector with support for multiple values.
+#'
+#' Supported values include:
+#'
+#' - "span-top": This operation spans empty cells with the first non-empty
+#' cell, applied column by column.
+#' - "center-hspan": Center the cells that are horizontally spanned.
+#' - "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+#' - "default-theme": Applies the theme set in `set_flextable_defaults(theme_fun = ...)` to the new header part.
#' @param split a regular expression (unless `fixed = TRUE`)
#' to use for splitting.
#' @param fixed logical. If TRUE match `split` exactly,
@@ -1105,11 +1100,12 @@ separate_header <- function(x,
for (j in seq_len(nrow(ref_list))) {
if (ref_list[j, 1]) {
to <- rle(ref_list[j, ])$lengths[1] + 1
-
- x <- merge_at(
- x = x, i = seq(1, to), j = j,
- part = "header"
- )
+ if (all(x$header$spans$rows[seq(1, to), j] %in% 1)) {#can be v-merged
+ x <- merge_at(
+ x = x, i = seq(1, to), j = j,
+ part = "header"
+ )
+ }
if ("bottom-vspan" %in% opts) {
x <- valign(
diff --git a/man/separate_header.Rd b/man/separate_header.Rd
index a318e409..cb4a86a1 100644
--- a/man/separate_header.Rd
+++ b/man/separate_header.Rd
@@ -2,7 +2,7 @@
% Please edit documentation in R/augment_rows.R
\name{separate_header}
\alias{separate_header}
-\title{Separate collapsed colnames into multiple rows}
+\title{Split column names using a separator into multiple rows}
\usage{
separate_header(
x,
@@ -14,21 +14,16 @@ separate_header(
\arguments{
\item{x}{a flextable object}
-\item{opts}{optional treatments to apply
-to the resulting header part as a character
-vector with multiple supported values.
+\item{opts}{Optional treatments to apply to the resulting header part.
+This should be a character vector with support for multiple values.
-The supported values are:
+Supported values include:
\itemize{
-\item "span-top": span empty cells with the
-first non empty cell, this operation is made
-column by column.
-\item "center-hspan": center the cells that are
-horizontally spanned.
-\item "bottom-vspan": bottom align the cells treated
-when "span-top" is applied.
-\item "default-theme": apply to the new header part
-the theme set in \code{set_flextable_defaults(theme_fun = ...)}.
+\item "span-top": This operation spans empty cells with the first non-empty
+cell, applied column by column.
+\item "center-hspan": Center the cells that are horizontally spanned.
+\item "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+\item "default-theme": Applies the theme set in \code{set_flextable_defaults(theme_fun = ...)} to the new header part.
}}
\item{split}{a regular expression (unless \code{fixed = TRUE})
@@ -38,9 +33,9 @@ to use for splitting.}
otherwise use regular expressions.}
}
\description{
-If your variable names contain
-multiple delimited labels, they will be separated
-and placed in their own rows.
+This function is used to separate and place individual
+labels in their own rows if your variable names contain multiple
+delimited labels.
\if{html}{\out{
#' }}
#' @param x a flextable object
-#' @param opts optional treatments to apply
-#' to the resulting header part as a character
-#' vector with multiple supported values.
-#'
-#' The supported values are:
-#'
-#' * "span-top": span empty cells with the
-#' first non empty cell, this operation is made
-#' column by column.
-#' * "center-hspan": center the cells that are
-#' horizontally spanned.
-#' * "bottom-vspan": bottom align the cells treated
-#' when "span-top" is applied.
-#' * "default-theme": apply to the new header part
-#' the theme set in `set_flextable_defaults(theme_fun = ...)`.
+#' @param opts Optional treatments to apply to the resulting header part.
+#' This should be a character vector with support for multiple values.
+#'
+#' Supported values include:
+#'
+#' - "span-top": This operation spans empty cells with the first non-empty
+#' cell, applied column by column.
+#' - "center-hspan": Center the cells that are horizontally spanned.
+#' - "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+#' - "default-theme": Applies the theme set in `set_flextable_defaults(theme_fun = ...)` to the new header part.
#' @param split a regular expression (unless `fixed = TRUE`)
#' to use for splitting.
#' @param fixed logical. If TRUE match `split` exactly,
@@ -1105,11 +1100,12 @@ separate_header <- function(x,
for (j in seq_len(nrow(ref_list))) {
if (ref_list[j, 1]) {
to <- rle(ref_list[j, ])$lengths[1] + 1
-
- x <- merge_at(
- x = x, i = seq(1, to), j = j,
- part = "header"
- )
+ if (all(x$header$spans$rows[seq(1, to), j] %in% 1)) {#can be v-merged
+ x <- merge_at(
+ x = x, i = seq(1, to), j = j,
+ part = "header"
+ )
+ }
if ("bottom-vspan" %in% opts) {
x <- valign(
diff --git a/man/separate_header.Rd b/man/separate_header.Rd
index a318e409..cb4a86a1 100644
--- a/man/separate_header.Rd
+++ b/man/separate_header.Rd
@@ -2,7 +2,7 @@
% Please edit documentation in R/augment_rows.R
\name{separate_header}
\alias{separate_header}
-\title{Separate collapsed colnames into multiple rows}
+\title{Split column names using a separator into multiple rows}
\usage{
separate_header(
x,
@@ -14,21 +14,16 @@ separate_header(
\arguments{
\item{x}{a flextable object}
-\item{opts}{optional treatments to apply
-to the resulting header part as a character
-vector with multiple supported values.
+\item{opts}{Optional treatments to apply to the resulting header part.
+This should be a character vector with support for multiple values.
-The supported values are:
+Supported values include:
\itemize{
-\item "span-top": span empty cells with the
-first non empty cell, this operation is made
-column by column.
-\item "center-hspan": center the cells that are
-horizontally spanned.
-\item "bottom-vspan": bottom align the cells treated
-when "span-top" is applied.
-\item "default-theme": apply to the new header part
-the theme set in \code{set_flextable_defaults(theme_fun = ...)}.
+\item "span-top": This operation spans empty cells with the first non-empty
+cell, applied column by column.
+\item "center-hspan": Center the cells that are horizontally spanned.
+\item "bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
+\item "default-theme": Applies the theme set in \code{set_flextable_defaults(theme_fun = ...)} to the new header part.
}}
\item{split}{a regular expression (unless \code{fixed = TRUE})
@@ -38,9 +33,9 @@ to use for splitting.}
otherwise use regular expressions.}
}
\description{
-If your variable names contain
-multiple delimited labels, they will be separated
-and placed in their own rows.
+This function is used to separate and place individual
+labels in their own rows if your variable names contain multiple
+delimited labels.
\if{html}{\out{
 }}
diff --git a/tests/testthat/test-merge.R b/tests/testthat/test-merge.R
index 3b45390d..c9dc90d5 100644
--- a/tests/testthat/test-merge.R
+++ b/tests/testthat/test-merge.R
@@ -55,3 +55,34 @@ test_that("merged cells can be un-merged", {
ft <- merge_none(ft)
expect_true(all(ft$body$spans$columns == 1))
})
+
+test_that("separate_header", {
+ x <- data.frame(
+ Species = as.factor(c("setosa", "versicolor", "virginica")),
+ Sepal.Length_mean_zzz = c(5.006, 5.936, 6.588),
+ Sepal.Length_sd = c(0.35249, 0.51617, 0.63588),
+ Sepal.Width_mean = c(3.428, 2.77, 2.974),
+ Sepal.Width_sd_sfsf_dsfsdf = c(0.37906, 0.3138, 0.3225),
+ Petal.Length_mean = c(1.462, 4.26, 5.552),
+ Petal.Length_sd = c(0.17366, 0.46991, 0.55189),
+ Petal.Width_mean = c(0.246, 1.326, 2.026),
+ Petal.Width_sd = c(0.10539, 0.19775, 0.27465)
+ )
+
+ ft_1 <- flextable(x)
+ ft_1 <- separate_header(x = ft_1,
+ opts = c("span-top", "bottom-vspan")
+ )
+ header_txt <- flextable:::fortify_run(ft_1) |>
+ subset(.part %in% "header")
+ expect_equal(
+ object = header_txt$txt,
+ expected =
+ c("Species", "Sepal", "Sepal", "Sepal", "Sepal", "Petal", "Petal",
+ "Petal", "Petal", "", "Length", "Length", "Width", "Width", "Length",
+ "Length", "Width", "Width", "", "mean", "sd", "mean", "sd", "mean",
+ "sd", "mean", "sd", "", "zzz", "", "", "sfsf", "", "", "", "",
+ "", "", "", "", "dsfsdf", "", "", "", "")
+ )
+
+})
}}
diff --git a/tests/testthat/test-merge.R b/tests/testthat/test-merge.R
index 3b45390d..c9dc90d5 100644
--- a/tests/testthat/test-merge.R
+++ b/tests/testthat/test-merge.R
@@ -55,3 +55,34 @@ test_that("merged cells can be un-merged", {
ft <- merge_none(ft)
expect_true(all(ft$body$spans$columns == 1))
})
+
+test_that("separate_header", {
+ x <- data.frame(
+ Species = as.factor(c("setosa", "versicolor", "virginica")),
+ Sepal.Length_mean_zzz = c(5.006, 5.936, 6.588),
+ Sepal.Length_sd = c(0.35249, 0.51617, 0.63588),
+ Sepal.Width_mean = c(3.428, 2.77, 2.974),
+ Sepal.Width_sd_sfsf_dsfsdf = c(0.37906, 0.3138, 0.3225),
+ Petal.Length_mean = c(1.462, 4.26, 5.552),
+ Petal.Length_sd = c(0.17366, 0.46991, 0.55189),
+ Petal.Width_mean = c(0.246, 1.326, 2.026),
+ Petal.Width_sd = c(0.10539, 0.19775, 0.27465)
+ )
+
+ ft_1 <- flextable(x)
+ ft_1 <- separate_header(x = ft_1,
+ opts = c("span-top", "bottom-vspan")
+ )
+ header_txt <- flextable:::fortify_run(ft_1) |>
+ subset(.part %in% "header")
+ expect_equal(
+ object = header_txt$txt,
+ expected =
+ c("Species", "Sepal", "Sepal", "Sepal", "Sepal", "Petal", "Petal",
+ "Petal", "Petal", "", "Length", "Length", "Width", "Width", "Length",
+ "Length", "Width", "Width", "", "mean", "sd", "mean", "sd", "mean",
+ "sd", "mean", "sd", "", "zzz", "", "", "sfsf", "", "", "", "",
+ "", "", "", "", "dsfsdf", "", "", "", "")
+ )
+
+})