See https://busco.ezlab.org/busco_userguide.html for BUSCO documentation
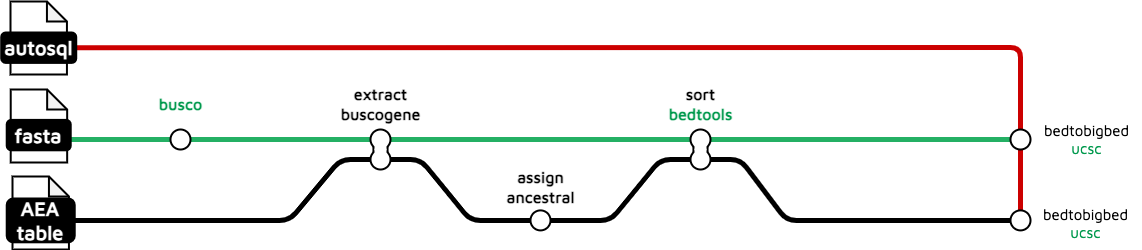 (from https://github.com/sanger-tol/treeval/blob/dev/docs/output.md#busco-analysis)
(from https://github.com/sanger-tol/treeval/blob/dev/docs/output.md#busco-analysis)
This has some odd DDL syntax, but we can probably figure it out with content expert help to explain, for example, why_ lepidoptera_ are a special case?
workflow BUSCO_ANNOTATION {
take:
dot_genome // channel: [val(meta), [ datafile ]]
reference_tuple // channel: [val(meta), [ datafile ]]
assembly_classT // channel: val(class)
lineageinfo // channel: val(lineage_db)
lineagespath // channel: val(/path/to/buscoDB)
buscogene_as // channel: val(dot_as location)
ancestral_table // channel: val(ancestral_table location)
main:
ch_versions = Channel.empty()
//
// MODULE: RUN BUSCO TO OBTAIN FULL_TABLE.CSV
// EMITS FULL_TABLE.CSV
//
BUSCO (
reference_tuple,
lineageinfo,
lineagespath,
[]
)
ch_versions = ch_versions.mix( BUSCO.out.versions.first() )
ch_grab = GrabFiles( BUSCO.out.busco_dir )
//
// MODULE: EXTRACT THE BUSCO GENES FOUND IN REFERENCE
//
EXTRACT_BUSCOGENE (
ch_grab
)
ch_versions = ch_versions.mix( EXTRACT_BUSCOGENE.out.versions )
//
// MODULE: SORT THE EXTRACTED BUSCO GENE
//
BEDTOOLS_SORT(
EXTRACT_BUSCOGENE.out.genefile,
[]
)
ch_versions = ch_versions.mix( BEDTOOLS_SORT.out.versions )
//
// MODULE: CONVERT THE BED TO BIGBED
//
UCSC_BEDTOBIGBED(
BEDTOOLS_SORT.out.sorted,
dot_genome.map{it[1]}, // Gets file from tuple (meta, file)
buscogene_as
)
ch_versions = ch_versions.mix( UCSC_BEDTOBIGBED.out.versions )
//
// LOGIC: AGGREGATE DATA AND SORT BRANCH ON CLASS
//
lineageinfo
.combine( BUSCO.out.busco_dir )
.combine( ancestral_table )
.branch {
lep: it[0].split('_')[0] == "lepidoptera"
general: it[0].split('_')[0] != "lepidoptera"
}
.set{ ch_busco_data }
//
// LOGIC: BUILD NEW INPUT CHANNEL FOR ANCESTRAL ID
//
ch_busco_data
.lep
.multiMap { lineage, meta, busco_dir, ancestral_table ->
busco_dir: tuple( meta, busco_dir )
atable: ancestral_table
}
.set{ ch_busco_lep_data }
//
// SUBWORKFLOW: RUN ANCESTRAL BUSCO ID (ONLY AVAILABLE FOR LEPIDOPTERA)
//
ANCESTRAL_GENE (
ch_busco_lep_data.busco_dir,
dot_genome,
buscogene_as,
ch_busco_lep_data.atable
)
ch_versions = ch_versions.mix( ANCESTRAL_GENE.out.versions )
emit:
ch_buscogene_bigbed = UCSC_BEDTOBIGBED.out.bigbed
ch_ancestral_bigbed = ANCESTRAL_GENE.out.ch_ancestral_bigbed
versions = ch_versions
}
First step needs a nextflow busco module, and that tool is available from the iuc in the Toolshed.
Next is EXTRACT_BUSCOGENE which runs another of the scripts in the /tree/bin directory,
get_busco_gene.sh $fulltable > ${prefix}_buscogene.csv
so need a new tool to run that. This tool in converting the busco table into BED to be consumed by JBrowse. Its a simple awk command, so I think we can convert this without a new tool.
Bedtools sort and ucsc_bedtobigbed are both used again so they will already be available.
Then there is some odd syntax involving an input database from the #1 yaml subworkflow. Looks like it is setting up a filesystem or streams for the #2 ANCESTRAL_GENE subworkflow described above. Hooboy. Will need a content expert to make sure the description being given here makes sense and that the data and parameters can be obtained correctly from the user.