-
Notifications
You must be signed in to change notification settings - Fork 1
/
Copy pathindex.qmd
496 lines (327 loc) · 15.4 KB
/
index.qmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
---
title: "R4CSR"
format: revealjs
editor: visual
---
# Book club
This is a slide deck summary of [R for Clinical Study Reporting and Submission](https://r4csr.org/)
There are three main parts of the book:
1. TLFs: a simulation of individual contributions
2. Clinical trial projects: a simulation of project leadership
3. eCTD submission: a simulation in package submission
In this summer book club we will get to just the first part, TLFs. [Join us!](https://datascience.arizona.edu/events/r-clinical-study-reporting)
## Get R help at UArizona
[Data & Visualization Drop-in](https://datascience.arizona.edu/events/data-viz-drop-0) hours \@ Main library
[Data Science Institute workshops](https://datascience.arizona.edu/calendar)
[CCT Data Science workshops](https://datascience.cct.arizona.edu/workshops)
## Preface
- R folder structure recommended
- [CDISC pilot](https://github.com/cdisc-org/sdtm-adam-pilot-project/tree/master/updated-pilot-submission-package/900172/m5/datasets/cdiscpilot01/analysis/adam/datasets)[data](https://github.com/elong0527/r4csr/tree/main/data-adam) is used throughout
- R packages needed
- not all packages are used in every chapter
```{r}
#| eval: false
#| echo: true
install.packages(c(
"dplyr" # Manipulate data
, "emmeans" # Least-square mean estimation
, "haven" # import SAS data
, "r2rtf" # Reporting in RTF format
, "survival" # kapplan meier curves
, "tidyr" # Manipulate data
, "table1" #Transfer data
))
```
## Living list of acronyms {.scrollable}
| Type | Acronym | Definition/Explanation |
|-------------------|-------------------|----------------------------------|
| clinical | CSR | Clinical study report |
| clinical | SDTM | standard data tabulation model |
| clinical | ADaM | Analysis dataset model |
| clinical | TLFs | Tables, listings, and figures |
| clinical | A&R | Analysis and reporting |
| clinical/computational | eCTD | Electronic common technical document |
| clinical | CDISC | Clinical Data Interchange Standards Consortium |
| clinical | ICH | International conference on harmonization |
| clinical | ICH E3 | ICH guidelines on structure and content of clinical study reports |
| computational | RTF | Rich text format |
| clinical | adae | Analysis dataset for adverse events |
| clinical/computational | ADSL | Subject-level analysis dataset |
| clinical/computational | ITT | intention to treat (i.e. none of the patients are excluded and the patients are analyzed according to the randomization scheme) |
| statistical | ANCOVA | Analysis of covariance |
| statistical | LOCF | last observation carried forward |
| statistical | K-M | Kaplan Meier curve/estimate |
| clinical | AEs | adverse events |
# Chapter 1
An overview of clinical study reports and the {r2rtf} package.
## Overview
- A CSR contains all methods and results of a study in \~16 sections
- TLFs are found in 10-12, 14, 16, +
- often word document format
- in this book we explore RTF
- The {r2rtf} package allows for the creation and export of publication quality tables and figures in rich text format
- Use the included `r2rtf_adae` dataset to explore functions in {r2rtf}
## r2rtf {#r2rtf}
After formatting your data as desired by {r2rtf}, a general workflow is as follows:
```{r}
#| echo: true
#| eval: false
head(tbl) %>%
rtf_body() %>% # Step 1 Add table attributes
rtf_encode() %>% # Step 2 Convert attributes to RTF encode
write_rtf("tlf/intro-ae1.rtf") # Step 3 Write to a .rtf file
```
Each table attribute is added individually, then the attributes are converted to RTF, and finally, you can export an object in RTF.
## Style with r2rtf
Use {r2rtf} to set specific design elements for your ouput:
```{r}
#| echo: true
#| eval: false
head(tbl) %>%
rtf_colheader(
colheader = " | Treatment",
col_rel_width = c(3, 6)
) %>%
rtf_colheader(
colheader = "Adverse Events | Placebo | Xanomeline High Dose | Xanomeline Low Dose",
border_top = c("", "single", "single", "single"),
col_rel_width = c(3, 2, 2, 2)
) %>%
rtf_body(col_rel_width = c(3, 2, 2, 2)) %>%
rtf_encode() %>%
write_rtf("tlf/intro-ae7.rtf")
```
A single {r2rtf} command may include columns, borders, headers, width designations and many other elements.
# Chapter 2
Section 10.1, Disposition of Participants
> The disposition of participants table reports the numbers of participants who were randomized, and who entered and completed each phase of the study, and the reasons for all post-randomization discontinuations, grouped by treatment and by major reason (lost to follow-up, adverse event, poor compliance, etc.) are reported.
## Disposition
**Step 1**: Count participants in the analysis population
**Step 2**: Calculate the number and percentage of participants who discontinued the study by treatment arm
**Step 3**: Calculate the numbers and percentages of participants who discontinued the study for different reasons, by treatment arm
**Step 4**: Calculate the number and percentage of participants who completed the study, by treatment arm
**Step 5**: Bind `n_rand`, `n_disc`, `n_reason`, and `n_complete` by row.
**Step 6+**: Write the final table to RTF
## Disposition of participants RTF
::: panel-tabset
### Code
```{r}
#| eval: false
#| echo: true
tbl_disp %>%
# Table title
rtf_title("Disposition of Participants") %>%
# First row of column header
rtf_colheader(" | Placebo | Xanomeline Low Dose| Xanomeline High Dose",
col_rel_width = c(3, rep(2, 3))
) %>%
# Second row of column header
rtf_colheader(" | n | (%) | n | (%) | n | (%)",
col_rel_width = c(3, rep(c(0.7, 1.3), 3)),
border_top = c("", rep("single", 6)),
border_left = c("single", rep(c("single", ""), 3))
) %>%
# Table body
rtf_body(
col_rel_width = c(3, rep(c(0.7, 1.3), 3)),
text_justification = c("l", rep("c", 6)),
border_left = c("single", rep(c("single", ""), 3))
) %>%
# Encoding RTF syntax
rtf_encode() %>%
# Save to a file
write_rtf("tlf/tbl_disp.rtf")
```
### Comments
With our bound data we follow our [workflow for {r2rtf}](#r2rtf): add attributes, convert to RTF, write out
- `|` separates every item, thus line 5 denotes 4 columns and line 9, 7 cols
- line 6 could also read\
`col_rel_width = c(3, 2, 2, 2)`
- and line 10 could also read\
`col_rel_width = c(3, 0.7, 1.3, 0.7, 1.3, 0.7, 1.3)`
:::
# Chapter 3
Section 11.1, Data Sets Analyzed
> The summary of analysis sets table reports on participants included in each efficacy analysis
## Writing functions
> "You should consider writing a function whenever you've copied and pasted a block of code more than twice"
>
> \- [R for Data Science](https://r4ds.had.co.nz/functions.html#when-should-you-write-a-function)
\
```{r}
#| eval: false
#| echo: true
fmt_num <- function(x, digits, width = digits + 4) {
formatC(x,
digits = digits,
format = "f",
width = width
)
}
```
## Summary of analysis sets
With helper functions `count_by` and `fmt_num` we can more easily prepare the dataset for a summary of analysis sets table with the following steps:\
\
**Step 1**: Bind the counts/percentages of the ITT population, the efficacy population, and the safety population by row using the `count_by()` function.\
**Step 2+**: Format the output from Step 2 using r2rtf.
# Chapter 4
Section 11.2 Demographic and other baseline characteristics
> Creating a simple table to summarize critical demographic and baseline characteristics of the participants
## R package {table1}
Efficiently summarizes this information and creates a HTML report\
\
\
```{r}
#| eval: false
#| echo: true
ana <- adsl %>%
mutate(
SEX = factor(SEX, c("F", "M"), c("Female", "Male")),
RACE = toTitleCase(tolower(RACE))
)
tbl <- table1(~ SEX + AGE + RACE | TRT01P, data = ana)
tbl
```
## Transferring the data
Transferring the output into a dataframe that contains only ASCII characters, recommended by regulatory agencies\
\
```{r}
#| eval: false
#| echo: true
tbl_base <- tbl %>%
as.data.frame() %>%
as_tibble() %>%
mutate(across(
everything(),
~ str_replace_all(.x, intToUtf8(160), " ")
))
names(tbl_base) <- str_replace_all(names(tbl_base), intToUtf8(160), " ")
tbl_base
```
## Final Formatting
Adjusting columns, headers, and indention.
```{r}
#| eval: false
#| echo: true
colheader1 <- paste(names(tbl_base), collapse = "|")
colheader2 <- paste(tbl_base[1, ], collapse = "|")
rel_width <- c(2.5, rep(1, 4))
tbl_base[-1, ] %>%
rtf_title(
"Baseline Characteristics of Participants",
"(All Participants Randomized)"
) %>%
rtf_colheader(colheader1,
col_rel_width = rel_width
) %>%
rtf_colheader(colheader2,
border_top = "",
col_rel_width = rel_width
) %>%
rtf_body(
col_rel_width = rel_width,
text_justification = c("l", rep("c", 4)),
text_indent_first = -240,
text_indent_left = 180
) %>%
rtf_encode() %>%
write_rtf("tlf/tlf_base.rtf")
```
## Summary of steps
**Step 1**: Use package {table1}
**Step 2**: Transfer the output from Step 1 into an ASCII data frame
**Step 3+**: Define the format of the RTF table
# Chapter 5
Section 11.4, Efficacy Results and Tabulations of Individual Participant
> Summarizing primary and secondary endpoints
## Efficacy Table
Combines two informative tables
1. Summary of observed data
- at baseline, week 24, and change from baseline
2. Pairwise comparisons with placebo
## Imputation {.center}
Missing data are inevitable
...why your data are missing can be highly informative
::: {style="color: red;"}
LOCF imputation is NOT a recommended imputation method
:::
Read more about the [prevention and treatment of missing data](https://www.ncbi.nlm.nih.gov/books/NBK209904/pdf/Bookshelf_NBK209904.pdf)
## ANCOVA
Analysis of covariance examines the realationship between an independent and dependent variable while controlling for a covariate
- Specifically, compare variance around means of different groups
Use {emmeans} to calculate within and between group least square means
## Efficacy tabulation steps
**Step 1**: Impute the missing values. In this example, we name the `ana` dataset after imputation as `ana_locf`.
**Step 2**: Calculate the mean and standard derivation of efficacy endpoint (i.e., `gluc`), and then format it into an RTF table.
**Step 3**: Calculate the pairwise comparison by ANCOVA model, and then format it into an RTF table.
**Step 4**: Combine the outputs from steps 4 and 5 by rows.
**Step 5+**: Format the output from Step 4 using r2rtf.
# Chapter 6
Section 11.4, Efficacy Results and Tabulations of Individual Participant
> The primary and secondary efficacy endpoints need to be summarized for each treatment group
## Survival Analysis
**aka time-to-event analysis**
\
A survival model explores the relationship between an outcome variable and a censored estimate of time to study dropout (for any number of reasons).
\
```{r}
#| label: kmExample
#| eval: false
#| echo: true
fit <- survfit(Surv(timeToEvent, 1 - censoredTime) ~ treatmentGroup
, data = clinicalData)
```
## Efficacy figure steps
**Step 1**: Define the analysis-ready dataset
**Step 2**: Save figures into `png` files
**Step 3+**: Create RTF output
# Chapter 7
Section 12.2 Adverse event (AEs) summary
> Summarize adverse eventing, the number of patients in each treatment group in whom the event occurred, and the rate of occurrence.
## Pivot wider
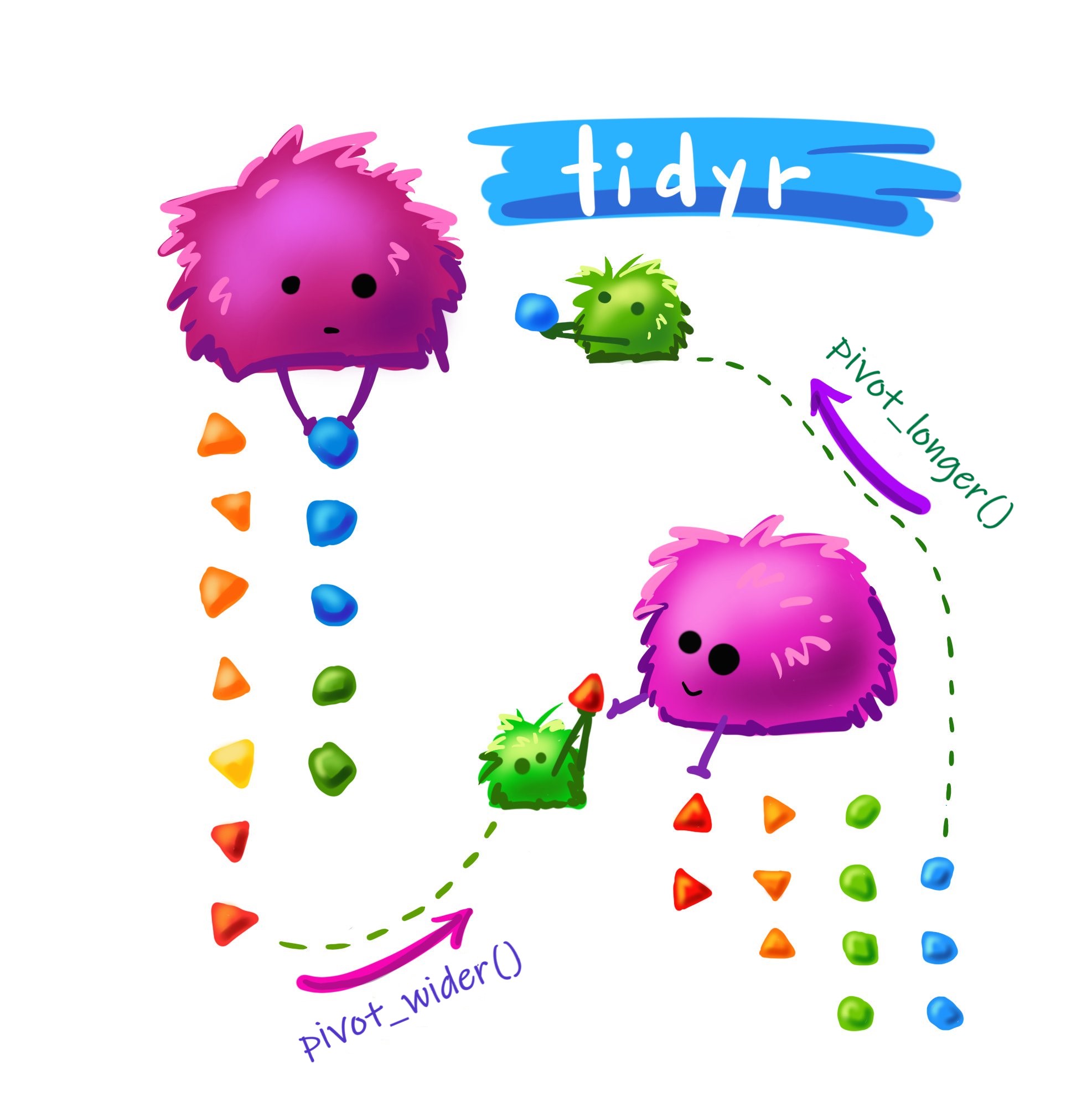{fig-alt="fuzzy monsters moving colorful rocks to demonstrate items in a dataframe moving between long and wide format" fig-align="center"}
## AE summary steps
**Step 1**: Summarize participants in population
**Step 2**: Summarize participants in population by AE criteria of interest
**Step 3**: Combine summaries
**Step 4+**: Format using r2rtf
# Chapter 8
Section 12.2 Specific AEs
> Report each adverse event, the number of patients in each treatment group in whom the event occurred, and the rate of occurrence.
## `page` functions
The AE table introduces us to two advanced table features:
- group content: AE can be summarized in multiple nested layers. (e.g., by system organ class (SOC, `AESOC`) and specific AE term (`AEDECOD`))
- `page_by()`
- pagenization: there are many AE terms that can not be covered in one page. Column headers and SOC information need to be repeated on every page.
- `pageby_row()`
## AE tabulation steps
**Step 1**: Count the number of participants by SOC and treatment arm
**Step 2**: Count the number of participants in each AE term by SOC, AE term, and treatment arm
**Step 3**: Count the number of participants in each arm
**Step 4**: Combine counts
**Step 5+**: Format the output by using r2rtf
# Chapter 9
All TLFs are then assembled by
1. Combining RTF source code in individual files into one large RTF file.
2. Leveraging the `Toggle Fields` feature in Microsoft Word to embed RTF files using hyperlinks.
## Combine source code
\
```{r}
#| label: assembleRTF1
#| eval: false
#| echo: true
tlf_path <- c(
"tlf/tbl_disp.rtf", # Disposition table
"tlf/tlf_eff.rtf", # Efficacy table
"tlf/tlf_km.rtf" # K-M plot
)
r2rtf::assemble_rtf(
input = tlf_path,
output = "tlf/rtf-combine.rtf"
)
```
## Leverage Microsoft Word
- Absolute paths
{fig-alt="folder icons in a directory tree with numerous examples of absolute (from root) vs relative (to another folder) paths"}
- Alt + F9 to "Toggle Fields"
# Thank you!!