From 3ea985f3e52d09e59ba843b84ab419ba3573b7e1 Mon Sep 17 00:00:00 2001
From: KianBV <91744358+KianBV@users.noreply.github.com>
Date: Tue, 8 Oct 2024 18:09:30 +0200
Subject: [PATCH] Update results.md
---
docs/project/results.md | 44 ++++++++++++++++++++++++++++++++++-------
1 file changed, 37 insertions(+), 7 deletions(-)
diff --git a/docs/project/results.md b/docs/project/results.md
index b6d3701..9f0cae7 100644
--- a/docs/project/results.md
+++ b/docs/project/results.md
@@ -12,6 +12,36 @@ The SIAH family recognizes its targets through a PXAXVXP degron motif [^E3_7], w
+## Selection of SIAH1/2 canonical targets
+
+The selection of canonical SIAH1/2 targets, needed to test the evolution logic, was performed based on the following criteria:
+
+1. The canonical substrate should be mainly cytoplasmic and not inserted or localised near the plasma membrane or in other cell organelles.
+2. The canonical Substrate should not natively form di- or other multi-mers as this could interfere with the selection system by tying two or more C-terminal RNAP subunits together.
+3. Post-translational modifications (PTMs) on the canonical substrate should not be necessary for its ubiquitination, as we cannot specifically control PTMs of substrates.
+4. The size of the canonical substrate should not exceed 300 amino acids in size, as this could also compromise our synthetic evolutionary system
+5. The structure of the protein and interaction with the selected E3 ligase (SIAH1/2) should be characterised.
+
+Based on these five criteria, we searched published ubiquitination databases (10.1093/database/baab010, 10.1093/nar/gkab962) for canonical substrates. EGLN1, EGLN3 and ɑ-Synuclein were selected as canonical substrates from the mentioned databases as they matched our criteria the best.
+
+| Canonical substrate | Protein size (aa) | Degron | Ubiquitination site | Canonical E3 ligase |
+| ------------- | ------------- | ------------- | ------------- | ------------- |
+| EGLN3 | 239 | 176-ADVEP-180 | K(159,172) | SIAH1/2 |
+| EGLN1 | 426 | 69-VGP-72, 376-VQP-379* | K256 | SIAH1/2 |
+| ɑ-Synuclein | 140 | 118-VDP-120 | K(6,10,12,21,23,32,34) | SIAH1/2 |
+
+* For EGLN1 accurate degron was not identified, though two VXP motifs exist of which at least one should act as SIAH1/2 degron.
+
+ɑ-Synuclein is well known for causing Parkinson's disease (PD) and multiple systems atrophy (MSA). Therefore, optimization of SIAH1/2 binding to ɑ-Synuclein could be an attractive alternative evolution strategy in case reprogramming SIAH1/2 specificity does not work as planned.
+
+NLRP3 was selected as an evolutionary target, for its mentioned clinical relevance in hepatic and neurodegenerative diseases. Furthermore, NLRP3 is an attractive target as it contains two VXP motifs (at positions 200 and 707), located in disordered regions on the surface of NLRP3 (Figure). This motif seems to be highly conserved among canonical SIAH1/2 substrates. Therefore, we assume our target choice is limited to proteins naturally displaying this motif on its surface, while other residues involved in binding could be changed. This would allow an evolution of SIAH1/2 to adapt for recognizing different VXP surrounding residues, while retaining the conserved VXP binding motif. Furthermore, lysins are found within the VXP surrounding residues, which are needed for ubiquitination. Remarkably, one of these lysins (K689) is natively polyubiquitinated and leads to the canonical degradation of NLRP3 (Liang, Damianou, Di Daniel, & Kessler, 2021), hence suggesting that polyubiquitination of K689 is a viable strategy of inducing NLRP3 degradation. With 1036 amino acids and its potential for oligomerization, NLRP3 could constrain our evolutionary system. Thus two peptide fragments of NLRP3 containing the VXP motif as well as surrounding residues were incorporated in the selection system.
+
+
+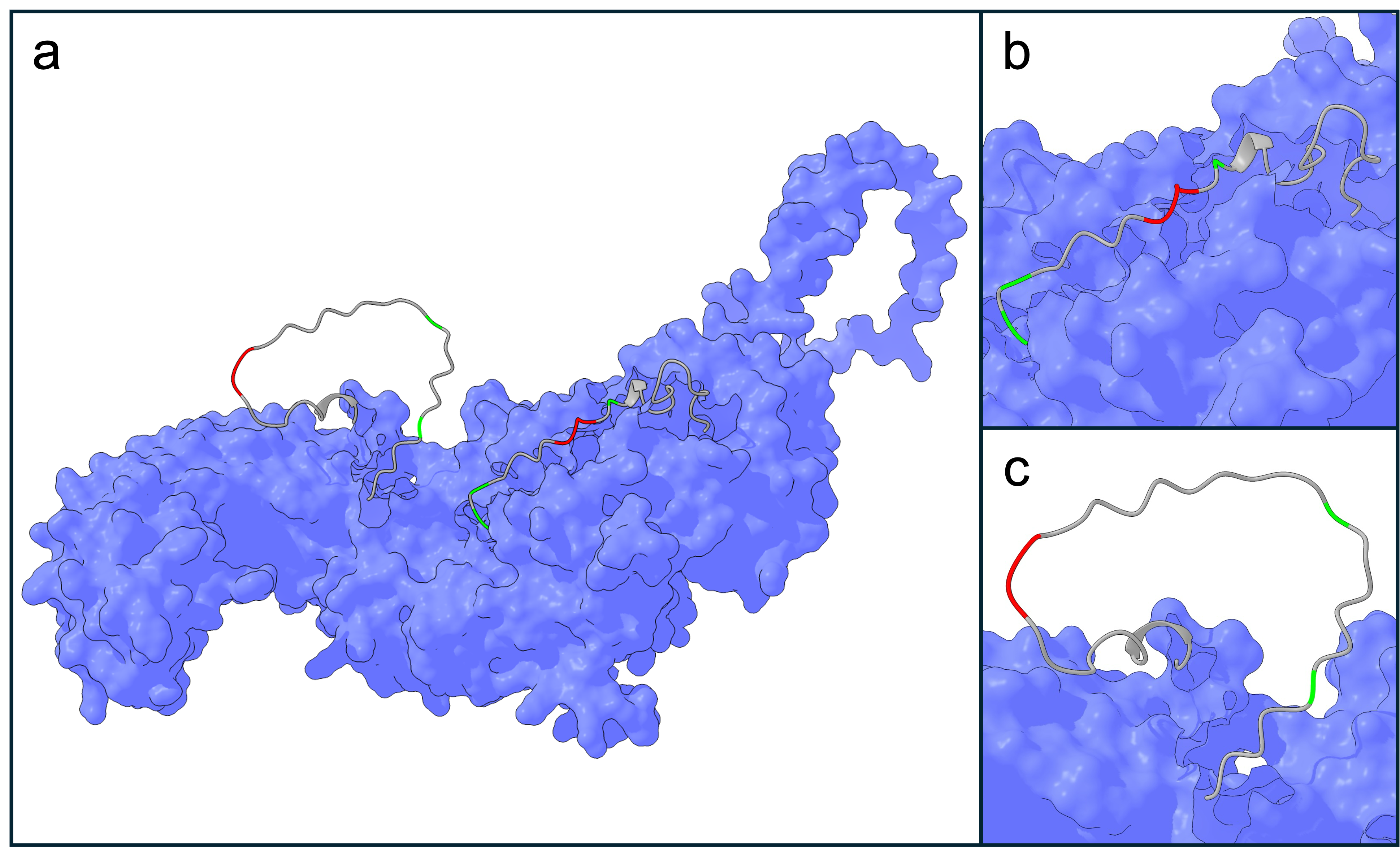
+ Figure 2: NLRP3 structure prediction by AlphaFold (AlphaFold protein structure database: AF-Q96P20-F1-v4). (a) NLRP3 contains two VXP motifs: (b) 200-VSP-203 and (c) 707-VLP-710, shown in red. Proximal lysins exist near the VXP degrons (green). Two short fragments, containing the VXP motif and surrounding lysins, were chosen as peptide substrates from NLRP3 for the evolutionary system (grey).
+
+
+
## Development of an E3 ligase PACE evolutionary system
### Ubiquitination-dependent selection logic
To evolve the SIAH1/2 E3 ubiquitin ligases using the PACE system, we needed to link the ligase’s activity directly to phage propagation. To achieve this, we utilised a modified T7 bacteriophage RNA polymerase (RNAP) that had been split into two halves. Normally, this split RNAP is inactive unless both halves are brought close together within the cell, forming a complete, functional complex. We designed a system where the RNAP halves would only assemble if the E3 ligase successfully ubiquitinated its target.
@@ -20,7 +50,7 @@ In this setup, one half of the RNAP is fused to ubiquitin, while the other half
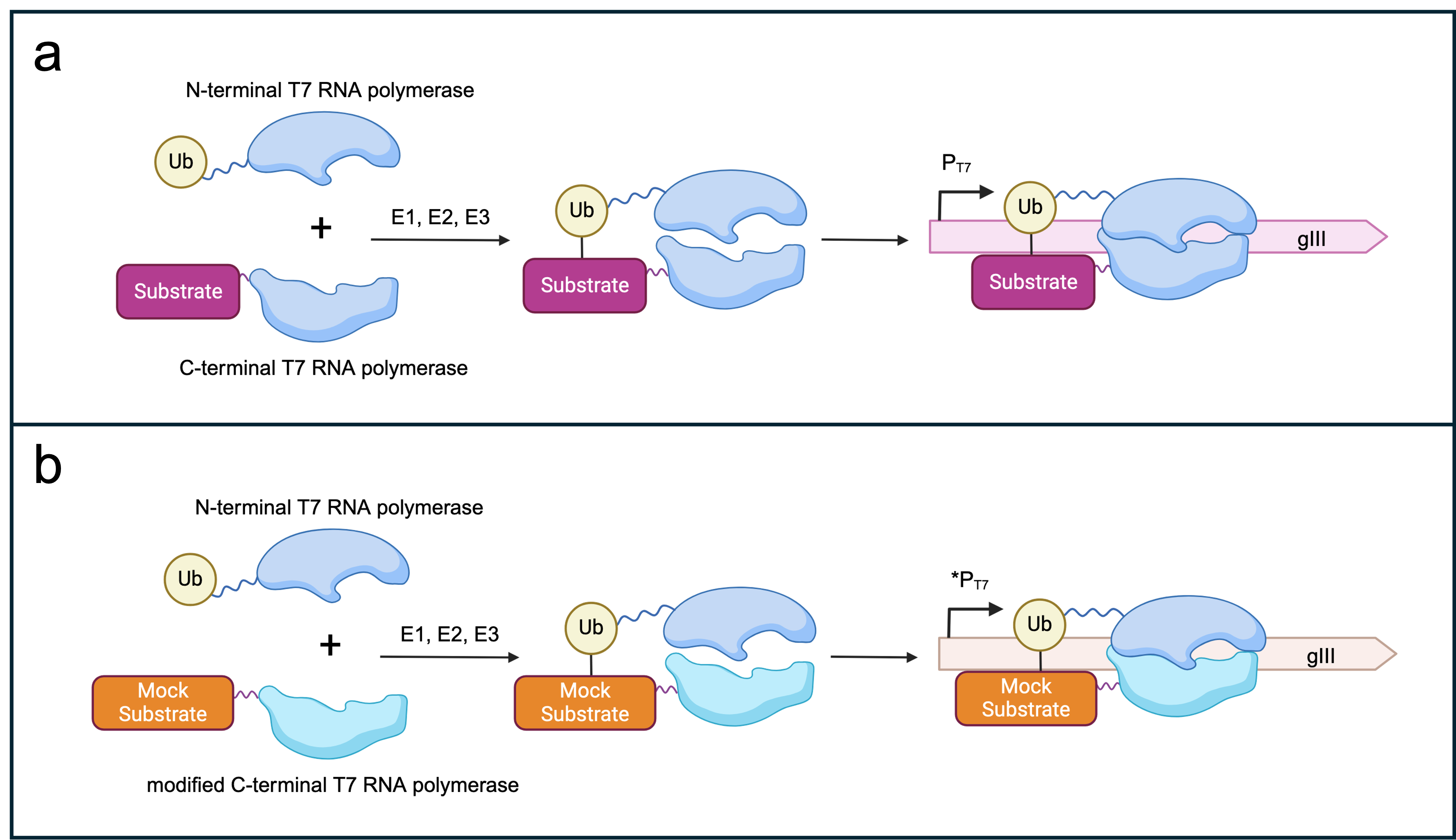
- Figure X: Selection logic for SIAH1/2-dependent _gIII_ expression. (a) Split T7 RNAP subunits fused to ubiquitin or a canonical substrate of SIAH1/2. The presence of E1, E2 and E3 (SIAH1/2) should lead to the assembly of the T7 RNAP subunits and thereby gIII transcription under the control of a T7 promoter. (b) Potential off-target effects of the evolved SIAH1/2 could be selected against by punishing spurious ubiquitination of a mock substrate by E3 ligase. In a new AP1neg plasmid, a mutated version of the C-term RNAP subunit that recognises a modified T7 promoter sequence [1] is fused to a mock substrate. A non-functional gIII (here, mock _gIII_) is placed under the control of the modified T7 promoter. Recognition and subsequent ubiquitination of the mock substrate by the evolved E3 ligase leads to the expression of mock _gIII_. Consequently, the phage offspring are not able to propagate further. Figure created with BioRender.com.
+ Figure 3: Selection logic for SIAH1/2-dependent _gIII_ expression. (a) Split T7 RNAP subunits fused to ubiquitin or a canonical substrate of SIAH1/2. The presence of E1, E2 and E3 (SIAH1/2) should lead to the assembly of the T7 RNAP subunits and thereby gIII transcription under the control of a T7 promoter. (b) Potential off-target effects of the evolved SIAH1/2 could be selected against by punishing spurious ubiquitination of a mock substrate by E3 ligase. In a new AP1neg plasmid, a mutated version of the C-term RNAP subunit that recognises a modified T7 promoter sequence [1] is fused to a mock substrate. A non-functional gIII (here, mock _gIII_) is placed under the control of the modified T7 promoter. Recognition and subsequent ubiquitination of the mock substrate by the evolved E3 ligase leads to the expression of mock _gIII_. Consequently, the phage offspring are not able to propagate further. Figure created with BioRender.com.
@@ -33,7 +63,7 @@ We plan to run this system in a bioreactor to create a continuous evolutionary e
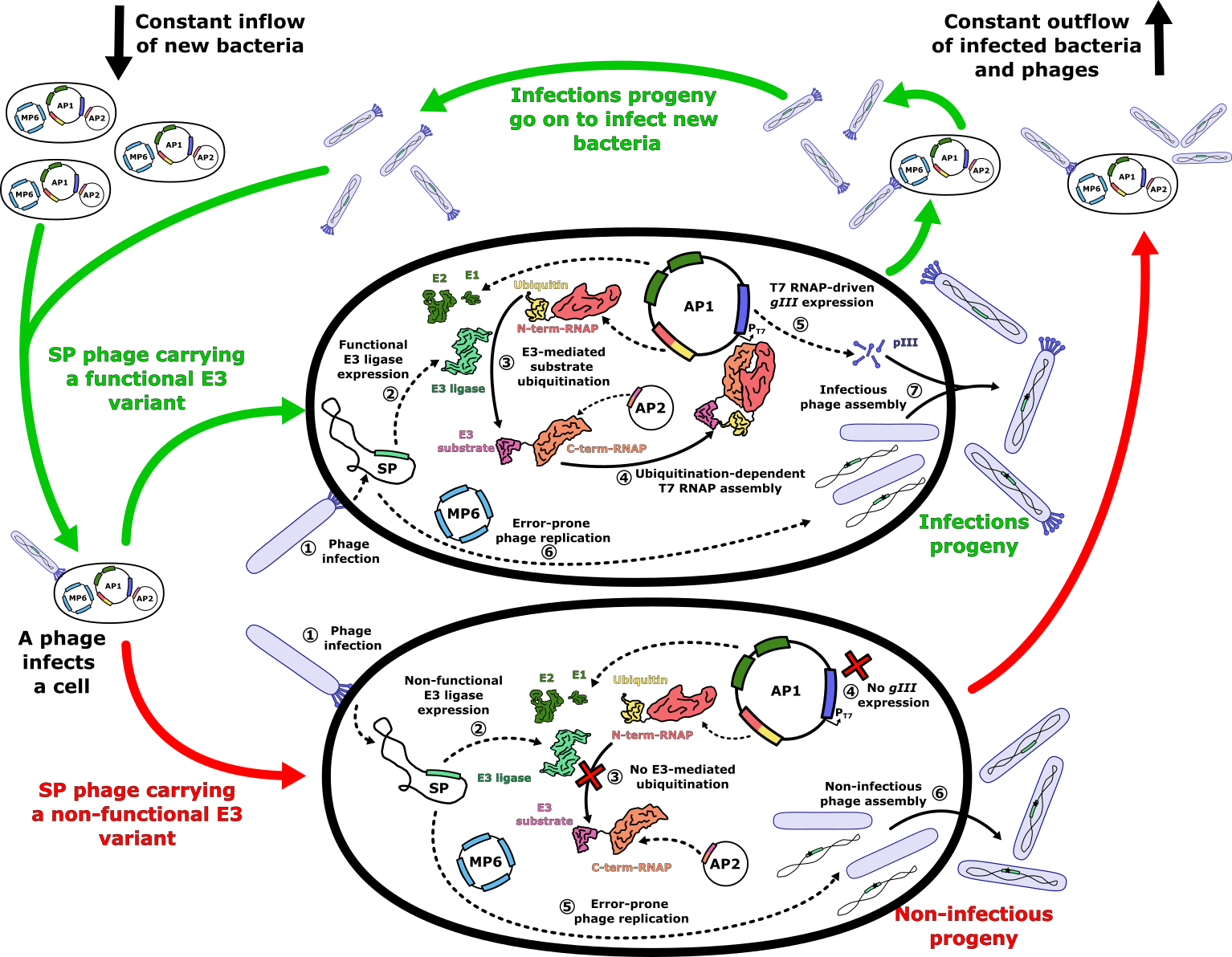
- Figure X: E3 ligase PACE evolutionary system. The PACE system operates within a lagoon with a constant inflow of new host cells and an outflow of phages and infected host cells. Upon infection of a host cell with a selection phage carrying a functional E3 ligase variant (green arrows), the E3 ligase ubiquitinates its target, which is fused to the C-terminal subunit of a split T7 RNA polymerase (RNAP), using ubiquitin fused to the N-terminal subunit of the split RNAP. The ubiquitin-mediated proximity of the split RNAP subunits assembles a functional T7 RNAP, which transcribes the _gIII_ gene required for the assembly of infectious progeny. In the case of a non-functional E3 variant (red arrows), the lack of assembled T7 RNAP prevents gIII expression, resulting in non-infectious phage progeny. During phage genome replication, the mutation plasmid MP6 increases the mutation rate, generating new E3 variants in the process. Infectious progeny carrying new E3 variants then go on to infect fresh host cells, repeating the cycle.
+ Figure 4: E3 ligase PACE evolutionary system. The PACE system operates within a lagoon with a constant inflow of new host cells and an outflow of phages and infected host cells. Upon infection of a host cell with a selection phage carrying a functional E3 ligase variant (green arrows), the E3 ligase ubiquitinates its target, which is fused to the C-terminal subunit of a split T7 RNA polymerase (RNAP), using ubiquitin fused to the N-terminal subunit of the split RNAP. The ubiquitin-mediated proximity of the split RNAP subunits assembles a functional T7 RNAP, which transcribes the _gIII_ gene required for the assembly of infectious progeny. In the case of a non-functional E3 variant (red arrows), the lack of assembled T7 RNAP prevents gIII expression, resulting in non-infectious phage progeny. During phage genome replication, the mutation plasmid MP6 increases the mutation rate, generating new E3 variants in the process. Infectious progeny carrying new E3 variants then go on to infect fresh host cells, repeating the cycle.
@@ -47,14 +77,14 @@ We tested if the E3 ligases SIAH1 and SIAH2 can trigger phage replication when a
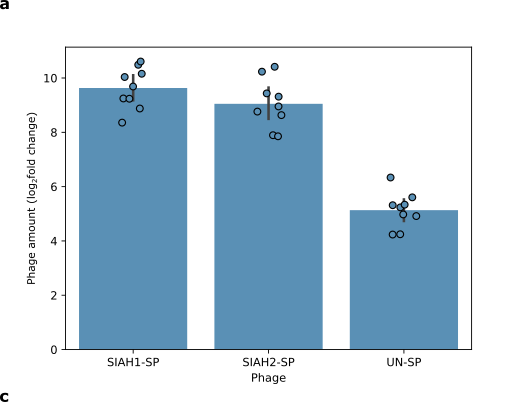
- Phages carrying SIAH1 or SIAH2 grow much better than phages carrying an unrelated protein. The fold change shows how much the number of phages increased or decreased (If the number of phages doubles, that is a two-fold increase).
+ Figure 5: Phages carrying SIAH1 or SIAH2 grow much better than phages carrying an unrelated protein. The fold change shows how much the number of phages increased or decreased (If the number of phages doubles, that is a two-fold increase).
We then looked at whether changing the target protein affects phage replication. We replaced EGLN3 (blue) with α-Synuclein (orange), which is also recognised by SIAH1/2. Changing the target protein changes the level of phage propagation. This shows that our system depends on SIAH1 or SIAH2 and the target protein interacting together.
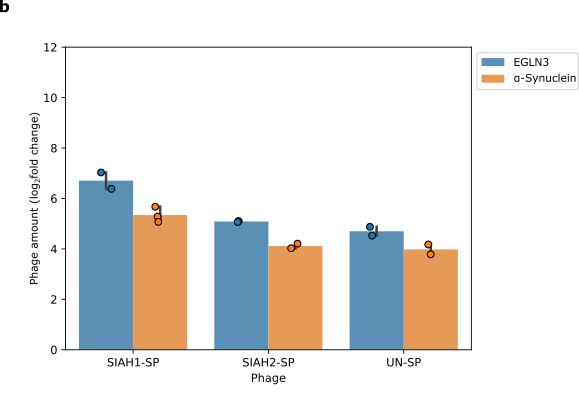
- Phage propagation is dependent on the substrate. The fold change shows how much the number of phages increased or decreased.
+ Figure 6: Phage propagation is dependent on the substrate. The fold change shows how much the number of phages increased or decreased.
@@ -62,7 +92,7 @@ Next, we looked at how changes in the degron would affect our system. We found t
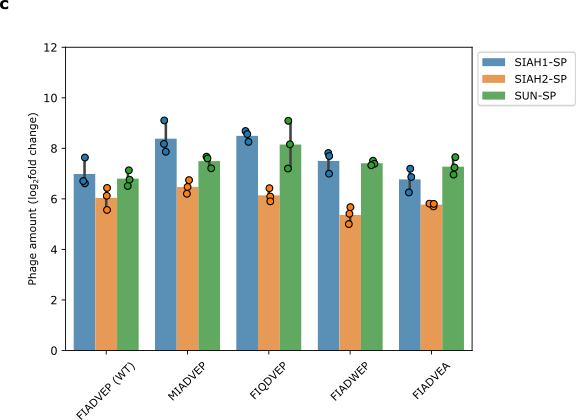
- Phage propagation is not affected by the disruption of the degron. The wildtype degron is: FIADVEP. The fold change shows how much the number of phages increased or decreased.
+ Figure 7: Phage propagation is not affected by the disruption of the degron. The wildtype degron is: FIADVEP. The fold change shows how much the number of phages increased or decreased.
We were able to show that phage propagation is dependent on both the substrate of the E3 ligase and the E3 ligase itself. However, we also observed a very high background phage propagation in our system. For our evolution to work, we need to be able to link phage proliferation to degron recognition, so this phage proliferation caused by other factors than the degron interferes with the use of this system for the directed evolution part of this project. The next step is to find out why this is happening and how we can modify our system so that it doesn't happen.
@@ -77,7 +107,7 @@ Both hypotheses lead to gIII expression independent of the E3 ligase activity. T
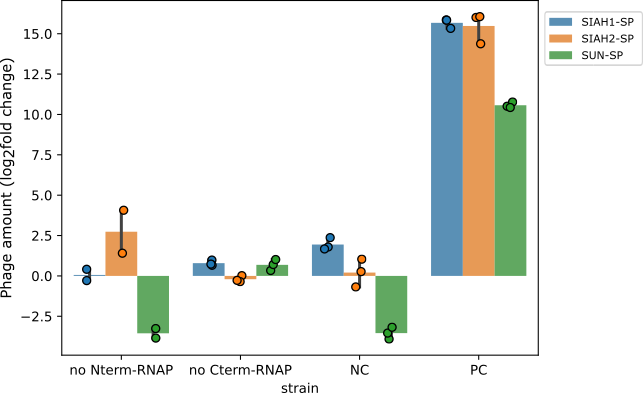
- Phage propagation in the presence and absence of split-RNAP components. This figure shows how much the phage population changed after overnight incubation with bacteria missing one of the two halves of the split-RNAP. The results are shown as a log2-fold change, meaning the numbers show how much the phage count increased or decreased, compared to the starting level. NC (negative control) is a bacterial strain lacking both parts of the system. PC (positive control) is a bacterial strain with the full system.
+ Figure 8: Phage propagation in the presence and absence of split-RNAP components. This figure shows how much the phage population changed after overnight incubation with bacteria missing one of the two halves of the split-RNAP. The results are shown as a log2-fold change, meaning the numbers show how much the phage count increased or decreased, compared to the starting level. NC (negative control) is a bacterial strain lacking both parts of the system. PC (positive control) is a bacterial strain with the full system.
### How can we reduce unwanted, E3-ligase independent page propagation?
@@ -91,7 +121,7 @@ To this end, we propagated the SIAH1 SP phages in bacterial cells that contain a
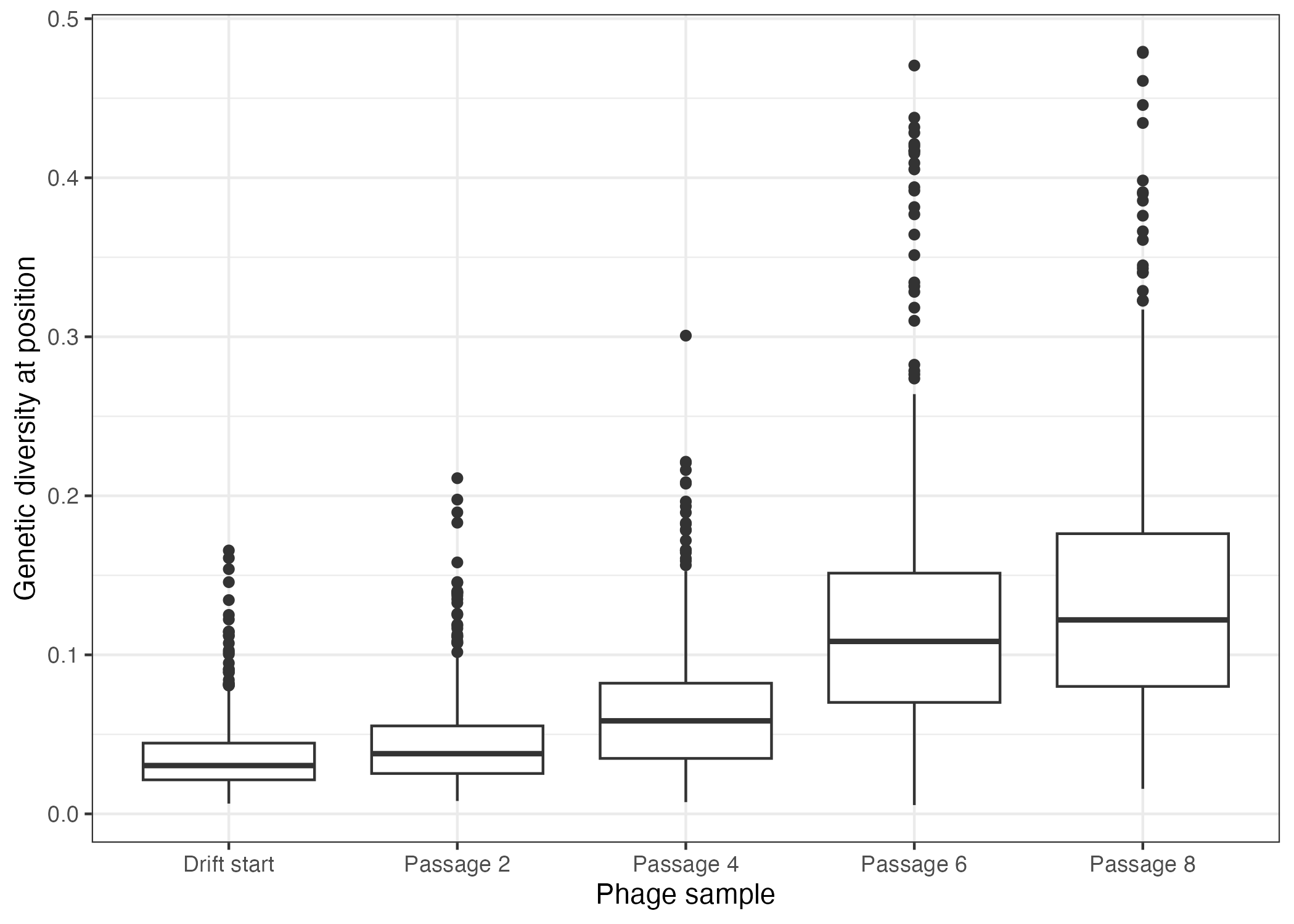
- Detection of mutated bases in SIAH1-SP during eight drift passages This figure illustrates the accumulation of mutations in the SIAH1-SP gene across eight cycles of drift. Each cycle represents one passage, during which the phages underwent random mutations in the absence of selection pressure. The data demonstrate how much genetic variation has been introduced over the course of the drift passages
+ Figure 9: Detection of mutated bases in SIAH1-SP during eight drift passages This figure illustrates the accumulation of mutations in the SIAH1-SP gene across eight cycles of drift. Each cycle represents one passage, during which the phages underwent random mutations in the absence of selection pressure. The data demonstrate how much genetic variation has been introduced over the course of the drift passages
## References