diff --git a/docs/project/results.md b/docs/project/results.md
index 0970883..22092ec 100644
--- a/docs/project/results.md
+++ b/docs/project/results.md
@@ -51,12 +51,12 @@ In this setup, one half of the RNAP is fused to ubiquitin, while the other half
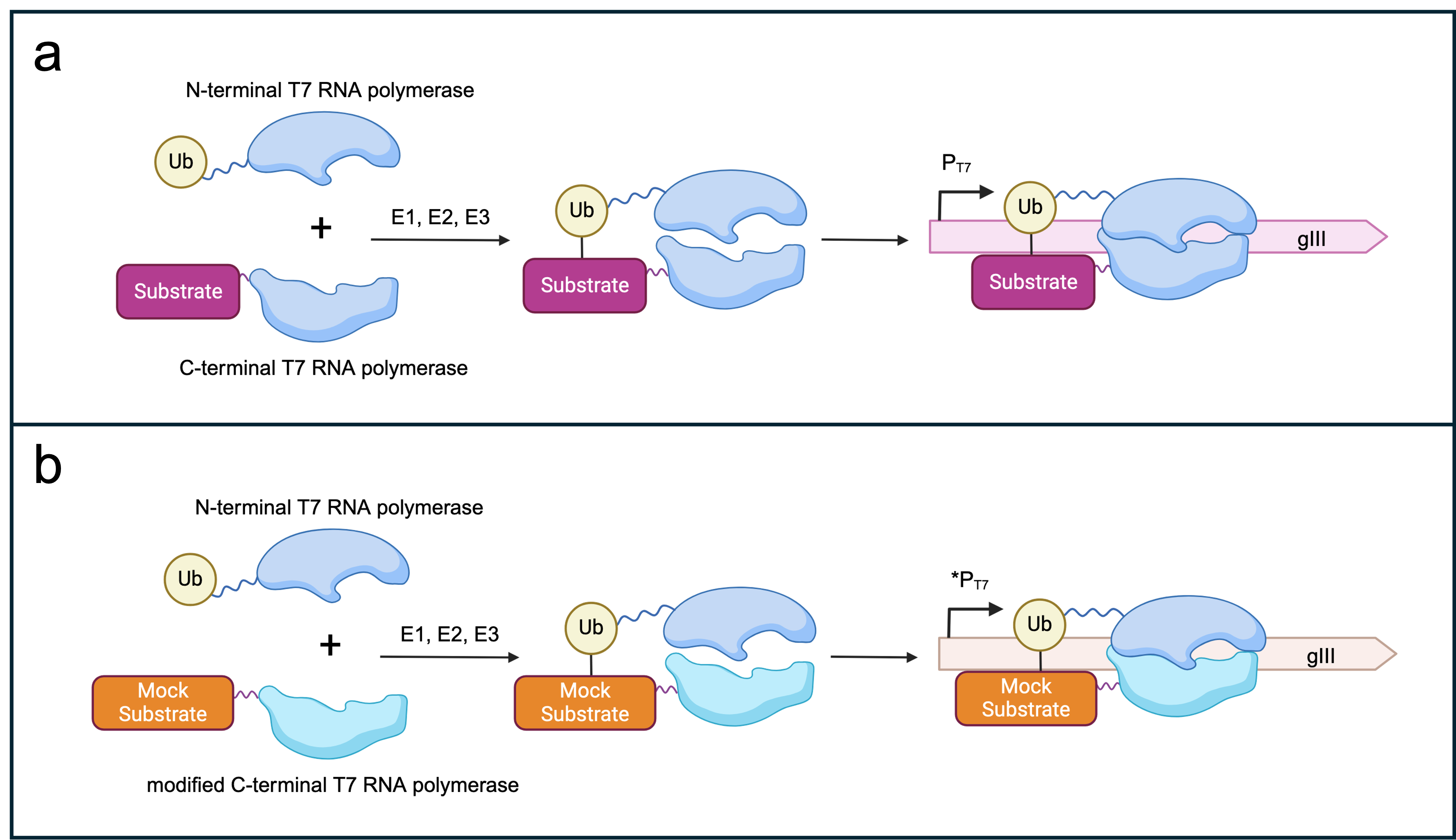
- Figure 3: Selection logic for SIAH1/2-dependent _gIII_ expression. (a) Split T7 RNAP subunits fused to ubiquitin or a canonical substrate of SIAH1/2. The presence of E1, E2 and E3 (SIAH1/2) should lead to the assembly of the T7 RNAP subunits and thereby gIII transcription under the control of a T7 promoter. (b) Potential off-target effects of the evolved SIAH1/2 could be selected against by punishing spurious ubiquitination of a mock substrate by E3 ligase. In a new AP1neg plasmid, a mutated version of the C-term RNAP subunit that recognises a modified T7 promoter sequence [1] is fused to a mock substrate. A non-functional gIII (here, mock _gIII_) is placed under the control of the modified T7 promoter. Recognition and subsequent ubiquitination of the mock substrate by the evolved E3 ligase leads to the expression of mock _gIII_. Consequently, the phage offspring are not able to propagate further. Figure created with BioRender.com.
+ Figure 3: Selection logic for SIAH1/2-dependent _gIII_ expression. (a) Split T7 RNAP subunits fused to ubiquitin or a canonical substrate of SIAH1/2. The presence of E1, E2 and E3 (SIAH1/2) should lead to the assembly of the T7 RNAP subunits and thereby _gIII_ transcription under the control of a T7 promoter. (b) Potential off-target effects of the evolved SIAH1/2 could be selected against by punishing spurious ubiquitination of a mock substrate by E3 ligase. In a new AP1neg plasmid, a mutated version of the C-term RNAP subunit that recognises a modified T7 promoter sequence [1] is fused to a mock substrate. A non-functional _gIII_ (here, mock _gIII_) is placed under the control of the modified T7 promoter. Recognition and subsequent ubiquitination of the mock substrate by the evolved E3 ligase leads to the expression of mock _gIII_. Consequently, the phage offspring are not able to propagate further. Figure created with BioRender.com.
### Assembling the PACE system
-To implement this system, we split the evolution process across three plasmids (Figure 4). The first plasmid is the selection phage (SP), which carries the SIAH1/2 gene and the phage genome but lacks the _gIII_, preventing the phage from propagating without the ligase’s activity. The second plasmid, accessory plasmid 1 (AP1), contains the genes for the E1 and E2 enzymes (which are required for ubiquitination but not normally present in bacteria), the N-terminal half of RNAP fused to ubiquitin, and the gIII controlled by the T7 promoter. The third plasmid, accessory plasmid 2 (AP2), contains the substrate fused to the C-terminal half of RNAP.
+To implement this system, we split the evolution process across three plasmids (Figure 4). The first plasmid is the selection phage (SP), which carries the SIAH1/2 gene and the phage genome but lacks the _gIII_, preventing the phage from propagating without the ligase’s activity. The second plasmid, accessory plasmid 1 (AP1), contains the genes for the E1 and E2 enzymes (which are required for ubiquitination but not normally present in bacteria), the N-terminal half of RNAP fused to ubiquitin, and the _gIII_ controlled by the T7 promoter. The third plasmid, accessory plasmid 2 (AP2), contains the substrate fused to the C-terminal half of RNAP.
This modular system allows us to easily swap out the substrate on AP2, enabling it to be applied to different E3 ligases or substrates. It also supports performing “substrate walks,” a process where we incrementally alter the amino-acid sequence of the substrate recognition motif to shift from a canonical target to a novel target of therapeutic interest. By doing this stepwise, we can control the selection pressure and gradually evolve SIAH1/2 to recognize new substrates.
@@ -64,7 +64,7 @@ We plan to run this system in a bioreactor to create a continuous evolutionary e
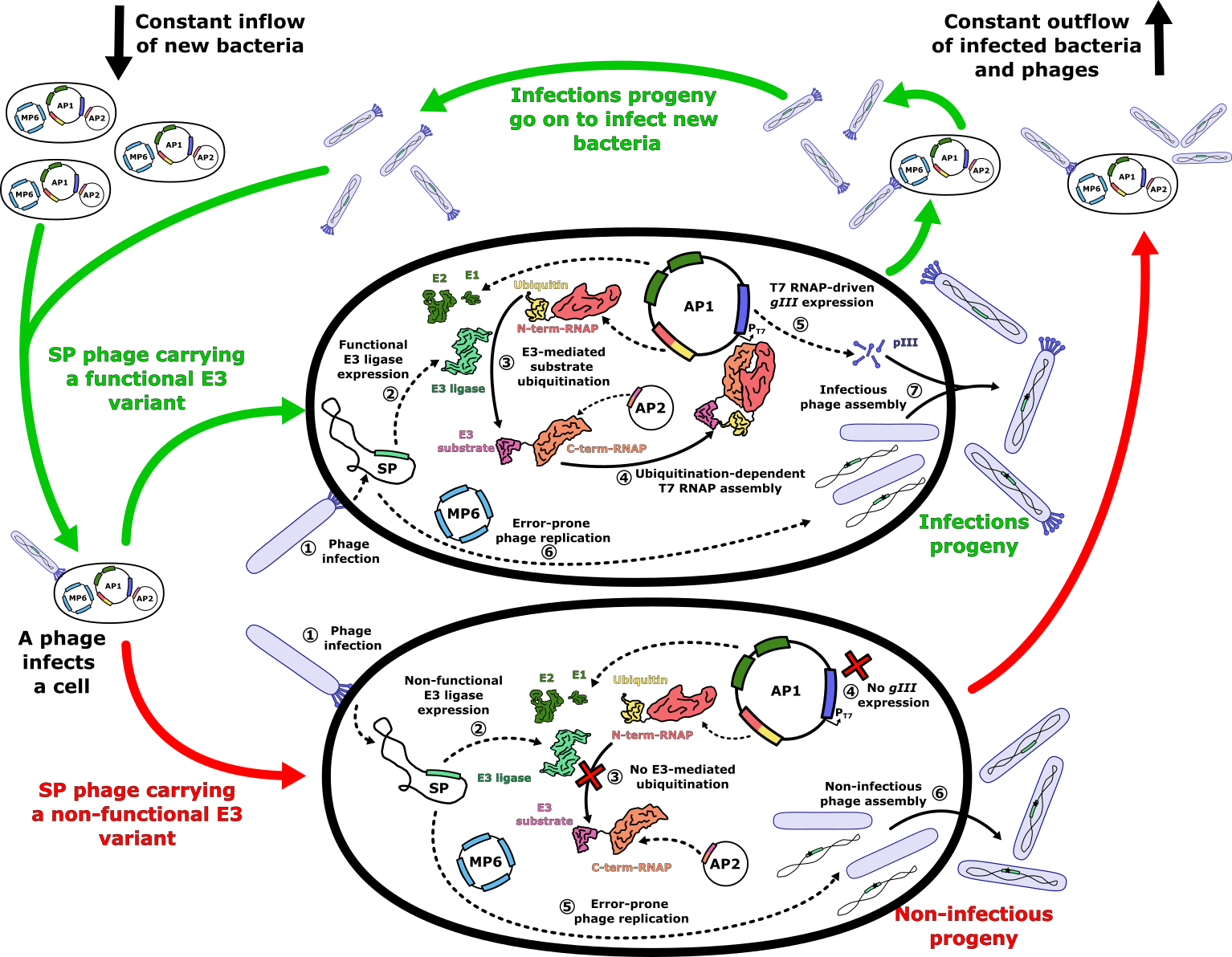
- Figure 4: E3 ligase PACE evolutionary system. The PACE system operates within a lagoon with a constant inflow of new host cells and an outflow of phages and infected host cells. Upon infection of a host cell with a selection phage carrying a functional E3 ligase variant (green arrows), the E3 ligase ubiquitinates its target, which is fused to the C-terminal subunit of a split T7 RNA polymerase (RNAP), using ubiquitin fused to the N-terminal subunit of the split RNAP. The ubiquitin-mediated proximity of the split RNAP subunits assembles a functional T7 RNAP, which transcribes the _gIII_ gene required for the assembly of infectious progeny. In the case of a non-functional E3 variant (red arrows), the lack of assembled T7 RNAP prevents gIII expression, resulting in non-infectious phage progeny. During phage genome replication, the mutation plasmid MP6 increases the mutation rate, generating new E3 variants in the process. Infectious progeny carrying new E3 variants then go on to infect fresh host cells, repeating the cycle.
+ Figure 4: E3 ligase PACE evolutionary system. The PACE system operates within a lagoon with a constant inflow of new host cells and an outflow of phages and infected host cells. Upon infection of a host cell with a selection phage carrying a functional E3 ligase variant (green arrows), the E3 ligase ubiquitinates its target, which is fused to the C-terminal subunit of a split T7 RNA polymerase (RNAP), using ubiquitin fused to the N-terminal subunit of the split RNAP. The ubiquitin-mediated proximity of the split RNAP subunits assembles a functional T7 RNAP, which transcribes the _gIII_ gene required for the assembly of infectious progeny. In the case of a non-functional E3 variant (red arrows), the lack of assembled T7 RNAP prevents _gIII_ expression, resulting in non-infectious phage progeny. During phage genome replication, the mutation plasmid MP6 increases the mutation rate, generating new E3 variants in the process. Infectious progeny carrying new E3 variants then go on to infect fresh host cells, repeating the cycle.
@@ -104,7 +104,7 @@ We suspected that the background phage propagation we observed could be caused b
1. Leaky transcription of _gIII_: the gene controlling phage growth is turned on without the split-RNAP components being present.
2. Spontaneous assembly of the split-RNAP subunits: The two halves of the RNAP are coming together on their own, without the need of ubiquitination of the target substrate.
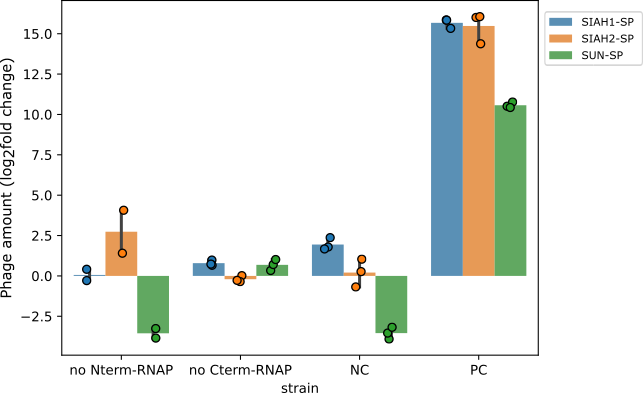
-Both hypotheses lead to _gIII_ expression independent of the E3 ligase activity. To test these hypotheses, we quantified phage propagation in cells that had only one half of the RNAP. In these cells, phage propagation was suppressed, confirming that both halves of the RNAP are required to activate gIII expression. This means that accidental activation of gIII wasn’t the issue. Instead, these results show that the two enzyme halves were probably joining together on their own, causing phage propagation without the involvement of the E3 ligase.
+Both hypotheses lead to _gIII_ expression independent of the E3 ligase activity. To test these hypotheses, we quantified phage propagation in cells that had only one half of the RNAP. In these cells, phage propagation was suppressed, confirming that both halves of the RNAP are required to activate _gIII_ expression. This means that accidental activation of _gIII_ wasn’t the issue. Instead, these results show that the two enzyme halves were probably joining together on their own, causing phage propagation without the involvement of the E3 ligase.