diff --git a/.circleci/config.yml b/.circleci/config.yml
index c96a86b28..88020d95d 100644
--- a/.circleci/config.yml
+++ b/.circleci/config.yml
@@ -33,7 +33,8 @@ jobs:
command: |
export PYTHONUNBUFFERED=1
# install dependencies and source code
- mamba install --verbose --yes gdal">=3" --file ${MINTPY_HOME}/requirements.txt
+ # pin gdal version because gdal-3.9 could not read the GMT-6 *.grd file properly
+ mamba install --verbose --yes --file ${MINTPY_HOME}/requirements.txt gdal"<3.9"
python -m pip install ${MINTPY_HOME}
# test installation
smallbaselineApp.py -h
@@ -49,31 +50,37 @@ jobs:
${MINTPY_HOME}/tests/dem_error.py
- run:
- name: Integration Test 1/4 - FernandinaSenDT128 (ISCE/topsStack)
+ name: Integration Test 1 - FernandinaSenDT128 (ISCE2/topsStack)
command: |
mkdir -p ${HOME}/data
${MINTPY_HOME}/tests/smallbaselineApp.py --dir ${HOME}/data --dset FernandinaSenDT128
- run:
- name: Integration Test 2/4 - SanFranSenDT42 (ARIA)
+ name: Integration Test 2 - SanFranSenDT42 (ARIA)
command: |
mkdir -p ${HOME}/data
${MINTPY_HOME}/tests/smallbaselineApp.py --dir ${HOME}/data --dset SanFranSenDT42
- run:
- name: Integration Test 3/5 - RidgecrestSenDT71 (HyP3)
+ name: Integration Test 3 - RidgecrestSenDT71 (ASF HyP3)
command: |
mkdir -p ${HOME}/data
${MINTPY_HOME}/tests/smallbaselineApp.py --dir ${HOME}/data --dset RidgecrestSenDT71
- run:
- name: Integration Test 4/5 - WellsEnvD2T399 (Gamma)
+ name: Integration Test 4 - SanFranBaySenD42 (GMTSAR)
+ command: |
+ mkdir -p ${HOME}/data
+ ${MINTPY_HOME}/tests/smallbaselineApp.py --dir ${HOME}/data --dset SanFranBaySenD42
+
+ - run:
+ name: Integration Test 5 - WellsEnvD2T399 (Gamma)
command: |
mkdir -p ${HOME}/data
${MINTPY_HOME}/tests/smallbaselineApp.py --dir ${HOME}/data --dset WellsEnvD2T399
- run:
- name: Integration Test 5/5 - WCapeSenAT29 (SNAP)
+ name: Integration Test 6 - WCapeSenAT29 (SNAP)
command: |
mkdir -p ${HOME}/data
${MINTPY_HOME}/tests/smallbaselineApp.py --dir ${HOME}/data --dset WCapeSenAT29
diff --git a/docs/README.md b/docs/README.md
index 79889a6e4..3a43e6f7d 100644
--- a/docs/README.md
+++ b/docs/README.md
@@ -51,7 +51,7 @@ smallbaselineApp.py ${MINTPY_HOME}/docs/templates/FernandinaSenDT128.txt
```
-  +
+ 
Results are plotted in **./pic** folder. To explore more data information and visualization, try the following scripts:
diff --git a/docs/demo_dataset.md b/docs/demo_dataset.md
index 788e8a3f3..321dc48d8 100644
--- a/docs/demo_dataset.md
+++ b/docs/demo_dataset.md
@@ -1,6 +1,6 @@
Here are example interferogram stacks pre-processed using different InSAR processors.
-### Sentinel-1 on Fernandina with ISCE ###
+### Sentinel-1 on Fernandina with ISCE2/topsStack ###
+ Area: Fernandina volcano at Galápagos Islands, Ecuador
+ Data: Sentinel-1 A/B descending track 128 during Dec 2014 - June 2018 (98 acquisitions; [Zenodo](https://zenodo.org/record/3952953))
@@ -14,7 +14,7 @@ smallbaselineApp.py ${MINTPY_HOME}/docs/templates/FernandinaSenDT128.txt
```
-  +
+ 
Relevant literature:
@@ -35,7 +35,41 @@ smallbaselineApp.py ${MINTPY_HOME}/docs/templates/SanFranSenDT42.txt
```
-  +
+  +
+
+
+Relevant literature:
+
++ Chaussard, E., R. Bürgmann, H. Fattahi, R. M. Nadeau, T. Taira, C. W. Johnson, and I. Johanson (2015), Potential for larger earthquakes in the East San Francisco Bay Area due to the direct connection between the Hayward and Calaveras Faults, _Geophysical Research Letters,_ 42(8), 2734-2741, doi:10.1002/2015GL063575.
+
+### Sentinel-1 of the 2019 Ridgecrest, California earthquake sequence with ASF HyP3 ###
+
++ Area: Owens Valley, California, USA ([USGS event page](https://earthquake.usgs.gov/earthquakes/eventpage/ci38457511/executive))
++ Data: Sentinel-1 descending track 71 during June - August 2019 (7 acquisitions; [Zenodo](https://zenodo.org/record/11049257))
++ Size: ~240 MB
+
+```bash
+wget https://zenodo.org/record/11049257/files/RidgecrestSenDT71.tar.xz
+tar -xvJf RidgecrestSenDT71.tar.xz
+cd RidgecrestSenDT71
+smallbaselineApp.py ${MINTPY_HOME}/docs/templates/RidgecrestSenDT71.txt
+```
+
+### Sentinel-1 on San Francisco Bay with GMTSAR ###
+
++ Area: San Francisco Bay, California, USA
++ Data: Sentinel-1 A/B descending track 42 during December 2014 - June 2024 (333 acquisitoins; [Zenodo](https://zenodo.org/records/12773014))
++ Size: ~2.3 GB
+
+```bash
+wget https://zenodo.org/records/12773014/files/SanFranBaySenD42.tar.xz
+tar -xvJf SanFranBaySenD42.tar.xz
+cd SanFranBaySenD42
+smallbaselineApp.py ${MINTPY_HOME}/docs/templates/SanFranBaySenD42.txt
+```
+
+
+ 
Relevant literature:
@@ -56,26 +90,13 @@ smallbaselineApp.py ${MINTPY_HOME}/docs/templates/WellsEnvD2T399.txt
```
- 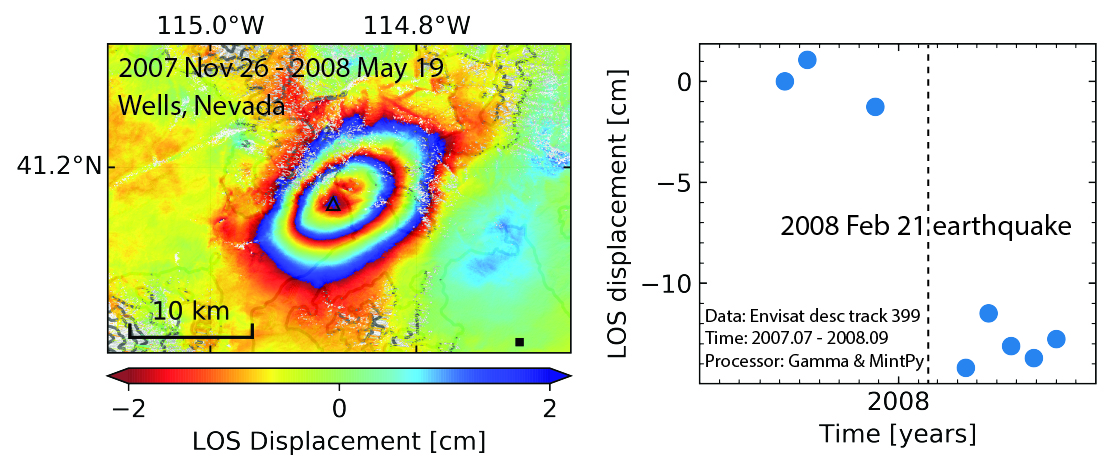 +
+ 
Relevant literature:
+ Nealy, J. L., H. M. Benz, G. P. Hayes, E. A. Bergman, and W. D. Barnhart (2017), The 2008 Wells, Nevada, Earthquake Sequence: Source Constraints Using Calibrated Multiple‐Event Relocation and InSAR, _Bulletin of the Seismological Society of America_, 107(3), 1107-1117, doi:10.1785/0120160298.
-### Sentinel-1 of the 2019 Ridgecrest, California earthquake sequence with HyP3 ###
-
-+ Area: Owens Valley, California, USA ([USGS event page](https://earthquake.usgs.gov/earthquakes/eventpage/ci38457511/executive))
-+ Data: Sentinel-1 descending track 71 during June - August 2019 (7 acquisitions; [Zenodo](https://zenodo.org/record/11049257))
-+ Size: ~240 MB
-
-```bash
-wget https://zenodo.org/record/11049257/files/RidgecrestSenDT71.tar.xz
-tar -xvJf RidgecrestSenDT71.tar.xz
-cd RidgecrestSenDT71
-smallbaselineApp.py ${MINTPY_HOME}/docs/templates/RidgecrestSenDT71.txt
-```
-
### Sentinel-1 on Western Cape, South Africa with SNAP ###
+ Area: West coast of Western Cape province, South Africa
@@ -103,7 +124,7 @@ smallbaselineApp.py ${MINTPY_HOME}/docs/templates/KujuAlosAT422F650.txt
```
-  +
+ 
Relevant literature:
diff --git a/docs/dir_structure.md b/docs/dir_structure.md
index 07c98b281..cc27c2a5c 100644
--- a/docs/dir_structure.md
+++ b/docs/dir_structure.md
@@ -463,9 +463,10 @@ $DATA_DIR/SanFranBaySenD42
├── baseline_table.dat
├── supermaster.PRM
├── geometry
-| ├── azimuth_angle.grd
| ├── dem.grd
-│ └── incidence_angle.grd
+| ├── azimuth_angle.grd
+│ ├── incidence_angle.grd
+│ └── water_mask.grd
├── interferograms
│ ├── 2004114_2004324
│ │ ├── corr_ll.grd
@@ -485,16 +486,17 @@ RLOOKS = 32 #[int], number of looks in the range direction
HEADING = -168.0 #[float], satellite heading angle, measured from the north in clockwise as positive
# One could open the *.kml file in Google Earth and measure it manually
-mintpy.load.processor = gmtsar
-mintpy.load.metaFile = $DATA_DIR/SanFranBaySenD42/supermaster.PRM
-mintpy.load.baselineDir = $DATA_DIR/SanFranBaySenD42/baseline_table.dat
+mintpy.load.processor = gmtsar
+mintpy.load.metaFile = $DATA_DIR/SanFranBaySenD42/supermaster.PRM
+mintpy.load.baselineDir = $DATA_DIR/SanFranBaySenD42/baseline_table.dat
##---------interferogram datasets:
-mintpy.load.unwFile = $DATA_DIR/SanFranBaySenD42/interferograms/*/unwrap_ll*.grd
-mintpy.load.corFile = $DATA_DIR/SanFranBaySenD42/interferograms/*/corr_ll*.grd
+mintpy.load.unwFile = $DATA_DIR/SanFranBaySenD42/interferograms/*/unwrap_ll*.grd
+mintpy.load.corFile = $DATA_DIR/SanFranBaySenD42/interferograms/*/corr_ll*.grd
##---------geometry datasets:
-mintpy.load.demFile = $DATA_DIR/SanFranBaySenD42/geometry/dem*.grd
-mintpy.load.incAngleFile = $DATA_DIR/SanFranBaySenD42/geometry/incidence_angle.grd
-mintpy.load.azAngleFile = $DATA_DIR/SanFranBaySenD42/geometry/azimuth_angle.grd
+mintpy.load.demFile = $DATA_DIR/SanFranBaySenD42/geometry/dem.grd
+mintpy.load.incAngleFile = $DATA_DIR/SanFranBaySenD42/geometry/incidence_angle.grd
+mintpy.load.azAngleFile = $DATA_DIR/SanFranBaySenD42/geometry/azimuth_angle.grd
+mintpy.load.waterMaskFile = $DATA_DIR/SanFranBaySenD42/geometry/water_mask.grd
```
### Gamma ###
diff --git a/docs/installation.md b/docs/installation.md
index a37e95186..5235eb26d 100644
--- a/docs/installation.md
+++ b/docs/installation.md
@@ -79,16 +79,18 @@ bash Miniforge3-Linux-x86_64.sh -b -p ~/tools/miniforge
Install dependencies into a new environment, e.g. named "insar":
```bash
-# Add "gdal'>=3'" below to install extra dependencies if you use ARIA, FRInGE, HyP3 or GMTSAR
-# Add "isce2" below to install extra dependencies if you use ISCE-2
+# Add "isce2" below to install extra dependencies if you use ISCE-2
+# Add "gdal" below to install extra dependencies if you use ARIA, FRInGE or HyP3
+# Add "gdal'<3.9'" below to install extra dependencies if you use GMTSAR
mamba create --name insar --file ~/tools/MintPy/requirements.txt
```
or install dependencies into an existing environment:
```bash
-# Add "gdal'>=3'" below to install extra dependencies if you use ARIA, FRInGE, HyP3 or GMTSAR
-# Add "isce2" below to install extra dependencies if you use ISCE-2
+# Add "isce2" below to install extra dependencies if you use ISCE-2
+# Add "gdal" below to install extra dependencies if you use ARIA, FRInGE or HyP3
+# Add "gdal'<3.9'" below to install extra dependencies if you use GMTSAR
mamba update --name my-existing-env --file ~/tools/MintPy/requirements.txt
```
diff --git a/docs/templates/SanFranBaySenD42.txt b/docs/templates/SanFranBaySenD42.txt
new file mode 100644
index 000000000..060120af8
--- /dev/null
+++ b/docs/templates/SanFranBaySenD42.txt
@@ -0,0 +1,28 @@
+# vim: set filetype=cfg:
+## manually add the following metadata
+HEADING = -168
+ALOOKS = 8
+RLOOKS = 32
+
+########## computing resource configuration
+mintpy.compute.maxMemory = 8
+mintpy.compute.cluster = local
+mintpy.compute.numWorker = 4
+
+########## 1. Load Data (--load to exit after this step)
+mintpy.load.processor = gmtsar #[isce, aria, hyp3, gmtsar, snap, gamma, roipac], auto for isce
+mintpy.load.metaFile = ../supermaster.PRM
+mintpy.load.baselineDir = ../baseline_table.dat
+##---------interferogram datasets:
+mintpy.load.unwFile = ../interferograms/*/unwrap_ll*.grd
+mintpy.load.corFile = ../interferograms/*/corr_ll*.grd
+##---------geometry datasets:
+mintpy.load.demFile = ../geometry/dem.grd
+mintpy.load.incAngleFile = ../geometry/incidence_angle.grd
+mintpy.load.azAngleFile = ../geometry/azimuth_angle.grd
+mintpy.load.waterMaskFile = ../geometry/water_mask.grd
+
+mintpy.reference.lalo = 37.69, -122.07
+mintpy.troposphericDelay.method = pyaps
+mintpy.deramp = no
+mintpy.topographicResidual = yes
diff --git a/src/mintpy/objects/stackDict.py b/src/mintpy/objects/stackDict.py
index 49158e40d..54c340f1d 100644
--- a/src/mintpy/objects/stackDict.py
+++ b/src/mintpy/objects/stackDict.py
@@ -695,10 +695,7 @@ def write2hdf5(self, outputFile='geometryRadar.h5', access_mode='w', box=None, x
c=str(compression)))
# read
- data = np.array(self.read(family=dsName,
- box=box,
- xstep=xstep,
- ystep=ystep)[0], dtype=dsDataType)
+ data = self.read(family=dsName, box=box, xstep=xstep, ystep=ystep)[0]
# water body: -1/True for water and 0/False for land
# water mask: 0/False for water and 1/True for land
@@ -708,6 +705,12 @@ def write2hdf5(self, outputFile='geometryRadar.h5', access_mode='w', box=None, x
print(' input file "{}" is water body (True/False for water/land), '
'convert to water mask (False/True for water/land).'.format(fname))
+ elif dsName == 'waterMask':
+ # GMTSAR water/land mask: 1 for land, and nan for water / no data
+ if np.sum(np.isnan(data)) > 0:
+ print(' convert NaN value for waterMask to zero.')
+ data[np.isnan(data)] = 0
+
elif dsName == 'height':
noDataValueDEM = -32768
if np.any(data == noDataValueDEM):
@@ -745,6 +748,7 @@ def write2hdf5(self, outputFile='geometryRadar.h5', access_mode='w', box=None, x
data = ut.wrap(data, wrap_range=[-180, 180]) # rewrap within -180 to 180
# write
+ data = np.array(data, dtype=dsDataType)
ds = f.create_dataset(dsName,
data=data,
chunks=True,
diff --git a/src/mintpy/prep_gmtsar.py b/src/mintpy/prep_gmtsar.py
index 3b8e230fa..1d31ad12c 100644
--- a/src/mintpy/prep_gmtsar.py
+++ b/src/mintpy/prep_gmtsar.py
@@ -20,6 +20,13 @@
#########################################################################
def get_ifg_file(ifg_dir, coord_type='geo', fbases=['corr', 'phase', 'phasefilt', 'unwrap']):
+ """Get the interferogram data file path for a given interferogram directory.
+
+ Parameters: ifg_dir - str, path of a given interferogram directory
+ coord_type - str, coordinate type in radar or geo
+ fbases - list(str), data file base name to look for
+ Returns: ifg_file - str, path of the interferogram data file
+ """
ifg_file = None
# search interferogram data file
@@ -40,7 +47,11 @@ def get_ifg_file(ifg_dir, coord_type='geo', fbases=['corr', 'phase', 'phasefilt'
def get_prm_files(ifg_dir):
- """Get the date1/2.prm files in the interferogram directory."""
+ """Get the date1/2.prm files in the interferogram directory.
+
+ Parameters: ifg_dir - str, path to the interferogram directory
+ Returns: prm_files - list(str), path to the PRM metadata files
+ """
prm_files = sorted(glob.glob(os.path.join(ifg_dir, '*.PRM')))
prm_files = [i for i in prm_files if os.path.splitext(os.path.basename(i))[0].isdigit()]
if len(prm_files) != 2:
@@ -49,7 +60,11 @@ def get_prm_files(ifg_dir):
def get_multilook_number(ifg_dir, fbases=['corr', 'phase', 'phasefilt', 'unwrap']):
- """Get the number of multilook in range and azimuth direction."""
+ """Get the number of multilook in range and azimuth direction.
+
+ Parameters: ifg_dir - str, path to the interferogram directory
+ Returns: a/rlooks - int, number of looks in the azimuth / range direction
+ """
# grab an arbitrary file in radar-coordiantes
rdr_file = get_ifg_file(ifg_dir, coord_type='radar')
@@ -62,7 +77,13 @@ def get_multilook_number(ifg_dir, fbases=['corr', 'phase', 'phasefilt', 'unwrap'
def get_lalo_ref(ifg_dir, prm_dict, fbases=['corr', 'phase', 'phasefilt', 'unwrap']):
- """Get the LAT/LON_REF1/2/3/4 from *_ll.grd file."""
+ """Get the LAT/LON_REF1/2/3/4 from *_ll.grd file.
+
+ Parameters: ifg_dir - str, path to the interferogram directory
+ prm_dict - dict, metadata from the PRM file
+ fbases - list(str), data file base name to look for
+ Returns: prm_dict - dict, metadata from the PRM file
+ """
# grab an arbitrary file in geo-coordiantes
geo_file = get_ifg_file(ifg_dir, coord_type='geo')
if not geo_file:
@@ -104,7 +125,13 @@ def get_lalo_ref(ifg_dir, prm_dict, fbases=['corr', 'phase', 'phasefilt', 'unwra
def get_slant_range_distance(ifg_dir, prm_dict, fbases=['corr', 'phase', 'phasefilt', 'unwrap']):
- """Get a constant slant range distance in the image center, for dataset in geo-coord."""
+ """Get a constant slant range distance in the image center, for dataset in geo-coord.
+
+ Parameters: ifg_dir - str, path to the interferogram directory
+ prm_dict - dict, metadata from the PRM file
+ fbases - list(str), data file base name to look for
+ Returns: prm_dict - dict, metadata from the PRM file
+ """
# grab an arbitrary file in geo/radar-coordiantes
geo_file = get_ifg_file(ifg_dir, coord_type='geo')
if not geo_file:
@@ -146,7 +173,14 @@ def read_baseline_table(fname):
#########################################################################
def extract_gmtsar_metadata(meta_file, unw_file, template_file, rsc_file=None, update_mode=True):
- """Extract metadata from GMTSAR interferogram stack."""
+ """Extract metadata from GMTSAR interferogram stack.
+
+ Parameters: meta_file - str, path to the reference metadata file in PRM format
+ unw_file - str, path to the unwrapped interferogram file
+ template_file - str, path to the template file
+ rsc_file - str, path to the output metadata file in RSC format
+ Returns: meta - dict, extract common metadata
+ """
# update_mode: check existing rsc_file
if update_mode and ut.run_or_skip(rsc_file, in_file=unw_file, readable=False) == 'skip':
@@ -161,11 +195,11 @@ def extract_gmtsar_metadata(meta_file, unw_file, template_file, rsc_file=None, u
# 2. read template file: HEADING, ORBIT_DIRECTION
template = readfile.read_template(template_file)
- for key in ['HEADING', 'ORBIT_DIRECTION', 'ALOOKS', 'RLOOKS']:
+ for key in ['HEADING', 'ALOOKS', 'RLOOKS']:
if key in template.keys():
meta[key] = template[key].lower()
else:
- raise ValueError('Attribute {} is missing! Please manually specify it in the template file.')
+ raise ValueError(f'Attribute {key} is missing! Please manually specify it in the template file.')
# 3. grab A/RLOOKS from radar-coord data file
#meta['ALOOKS'], meta['RLOOKS'] = get_multilook_number(ifg_dir)
@@ -213,8 +247,19 @@ def extract_gmtsar_metadata(meta_file, unw_file, template_file, rsc_file=None, u
return meta
-def prepare_geometry(geom_files, meta, update_mode=True):
- """Prepare .rsc file for all geometry files."""
+def prepare_geometry(template, meta, update_mode=True):
+ """Prepare .rsc file for all geometry files.
+
+ Parameters: template - dict, input file path/pattern
+ meta - dict, common metadata for the entire stack
+ """
+ # grab all specified geometry files
+ geom_files = []
+ key_prefix = 'mintpy.load.'
+ for key in ['demFile', 'lookupYFile', 'lookupXFile', 'incAngleFile',
+ 'azAngleFile', 'shadowMaskFile', 'waterMaskFile']:
+ if key_prefix + key in template.keys() and template[key_prefix + key]:
+ geom_files.append(glob.glob(template[key_prefix + key])[0])
num_file = len(geom_files)
if num_file == 0:
raise FileNotFoundError('NO geometry file found!')
@@ -242,7 +287,12 @@ def prepare_geometry(geom_files, meta, update_mode=True):
def prepare_stack(unw_files, meta, bperp_dict, update_mode=True):
- """Prepare .rsc file for all unwrapped interferogram files."""
+ """Prepare .rsc file for all unwrapped interferogram files.
+
+ Parameters: unw_files - list(str), path to the unwrapped interferogram files
+ meta - dict, common metadata for the entire stack
+ bperp_dict - dict, perpendicular baseline time series
+ """
num_file = len(unw_files)
if num_file == 0:
raise FileNotFoundError('NO unwrapped interferogram file found!')
@@ -288,13 +338,12 @@ def prepare_stack(unw_files, meta, bperp_dict, update_mode=True):
def prep_gmtsar(inps):
# read file path from template file
template = readfile.read_template(inps.template_file)
- inps.meta_file = glob.glob(template['mintpy.load.metaFile'])[0]
- inps.bperp_file = glob.glob(template['mintpy.load.baselineDir'])[0]
+ template = ut.check_template_auto_value(template)
+
+ inps.meta_file = glob.glob(template['mintpy.load.metaFile'])[0]
+ inps.bperp_file = glob.glob(template['mintpy.load.baselineDir'])[0]
inps.unw_files = sorted(glob.glob(template['mintpy.load.unwFile']))
inps.cor_files = sorted(glob.glob(template['mintpy.load.corFile']))
- inps.dem_file = glob.glob(template['mintpy.load.demFile'])[0]
- inps.inc_angle_file = glob.glob(template['mintpy.load.incAngleFile'])[0]
- inps.az_angle_file = glob.glob(template['mintpy.load.azAngleFile'])[0]
# extract common metadata
rsc_file = os.path.join(os.path.dirname(inps.meta_file), 'data.rsc')
@@ -311,7 +360,7 @@ def prep_gmtsar(inps):
# prepare metadata for geometry files
prepare_geometry(
- geom_files=[inps.dem_file, inps.inc_angle_file, inps.az_angle_file],
+ template=template,
meta=meta,
update_mode=inps.update_mode,
)
diff --git a/src/mintpy/utils/readfile.py b/src/mintpy/utils/readfile.py
index 9ef748984..46ad31612 100644
--- a/src/mintpy/utils/readfile.py
+++ b/src/mintpy/utils/readfile.py
@@ -1811,7 +1811,7 @@ def _attribute_gmtsar2roipac(prm_dict_in):
prm_dict['ANTENNA_SIDE'] = '1'
# orbdir -> ORBIT_DIRECTION
- key = 'obsdir'
+ key = 'orbdir'
if key in prm_dict_in.keys():
prm_dict['ORBIT_DIRECTION'] = {
'A' : 'ASCENDING',
diff --git a/tests/configs/SanFranBaySenD42.txt b/tests/configs/SanFranBaySenD42.txt
new file mode 100644
index 000000000..ef68e9f52
--- /dev/null
+++ b/tests/configs/SanFranBaySenD42.txt
@@ -0,0 +1,27 @@
+# vim: set filetype=cfg:
+## manually add the following metadata
+HEADING = -168
+ALOOKS = 8
+RLOOKS = 32
+
+########## computing resource configuration
+mintpy.compute.cluster = local
+
+########## 1. Load Data (--load to exit after this step)
+mintpy.load.processor = gmtsar #[isce, aria, hyp3, gmtsar, snap, gamma, roipac], auto for isce
+mintpy.load.metaFile = ../supermaster.PRM
+mintpy.load.baselineDir = ../baseline_table.dat
+##---------interferogram datasets:
+mintpy.load.unwFile = ../interferograms/*/unwrap_ll*.grd
+mintpy.load.corFile = ../interferograms/*/corr_ll*.grd
+##---------geometry datasets:
+mintpy.load.demFile = ../geometry/dem.grd
+mintpy.load.incAngleFile = ../geometry/incidence_angle.grd
+mintpy.load.azAngleFile = ../geometry/azimuth_angle.grd
+mintpy.load.waterMaskFile = ../geometry/water_mask.grd
+
+mintpy.reference.lalo = 37.69, -122.07
+mintpy.networkInversion.weightFunc = no # fast but not the best
+mintpy.deramp = no
+mintpy.topographicResidual = yes
+mintpy.topographicResidual.pixelwiseGeometry = no # fast but not the best
diff --git a/tests/requirements.txt b/tests/requirements.txt
index ef749423f..9815afdca 100644
--- a/tests/requirements.txt
+++ b/tests/requirements.txt
@@ -1,4 +1,4 @@
-gdal>=3 # for ISCE-2/3, ARIA, FRInGE, HyP3, GMTSAR users
+gdal<3.9 # for ISCE-2/3, ARIA, FRInGE, HyP3, GMTSAR users, gdal-3.9 could not read GMT *.grd file's coordinate info properly
isce2 # for ISCE-2 users
pre-commit # for developers
pyfftw
diff --git a/tests/smallbaselineApp.py b/tests/smallbaselineApp.py
index fb4c4d47a..0c212d7dc 100755
--- a/tests/smallbaselineApp.py
+++ b/tests/smallbaselineApp.py
@@ -26,6 +26,7 @@
'https://zenodo.org/record/6662079/files/SanFranSenDT42.tar.xz', # 280 MB
'https://zenodo.org/record/3952950/files/WellsEnvD2T399.tar.xz', # 280 MB
'https://zenodo.org/record/11049257/files/RidgecrestSenDT71.tar.xz', # 240 MB
+ 'https://zenodo.org/record/12772940/files/SanFranBaySenD42.tar.xz', # 290 MB
'https://zenodo.org/record/6661536/files/WCapeSenAT29.tar.xz', # 240 MB
'https://zenodo.org/record/3952917/files/KujuAlosAT422F650.tar.xz', # 230 MB
]
@@ -40,6 +41,7 @@
SanFranSenDT42 for ARIA
WellsEnvD2T399 for GAMMA
RidgecrestSenDT71 for HyP3
+ SanFranBaySenD42 for GMTSAR
WCapeSenAT29 for SNAP
KujuAlosAT422F650 for ROI_PAC
"""
 +
+ 
 +
+ 
 +
+ 
 +
+  +
+
 +
+ 
 +
+ 