diff --git a/assets/explore_high_dimensional_data.gif b/assets/explore_high_dimensional_data.gif
new file mode 100644
index 0000000..74f2a66
Binary files /dev/null and b/assets/explore_high_dimensional_data.gif differ
diff --git a/explore_high_dimensional_data/README.md b/explore_high_dimensional_data/README.md
new file mode 100644
index 0000000..8ce5662
--- /dev/null
+++ b/explore_high_dimensional_data/README.md
@@ -0,0 +1,43 @@
+# Explore high dimensional data
+
+[](https://marimo.app/github.com/marimo-team/examples/blob/main/explore_high_dimensional_data/explore_high_dimensional_data.py)
+
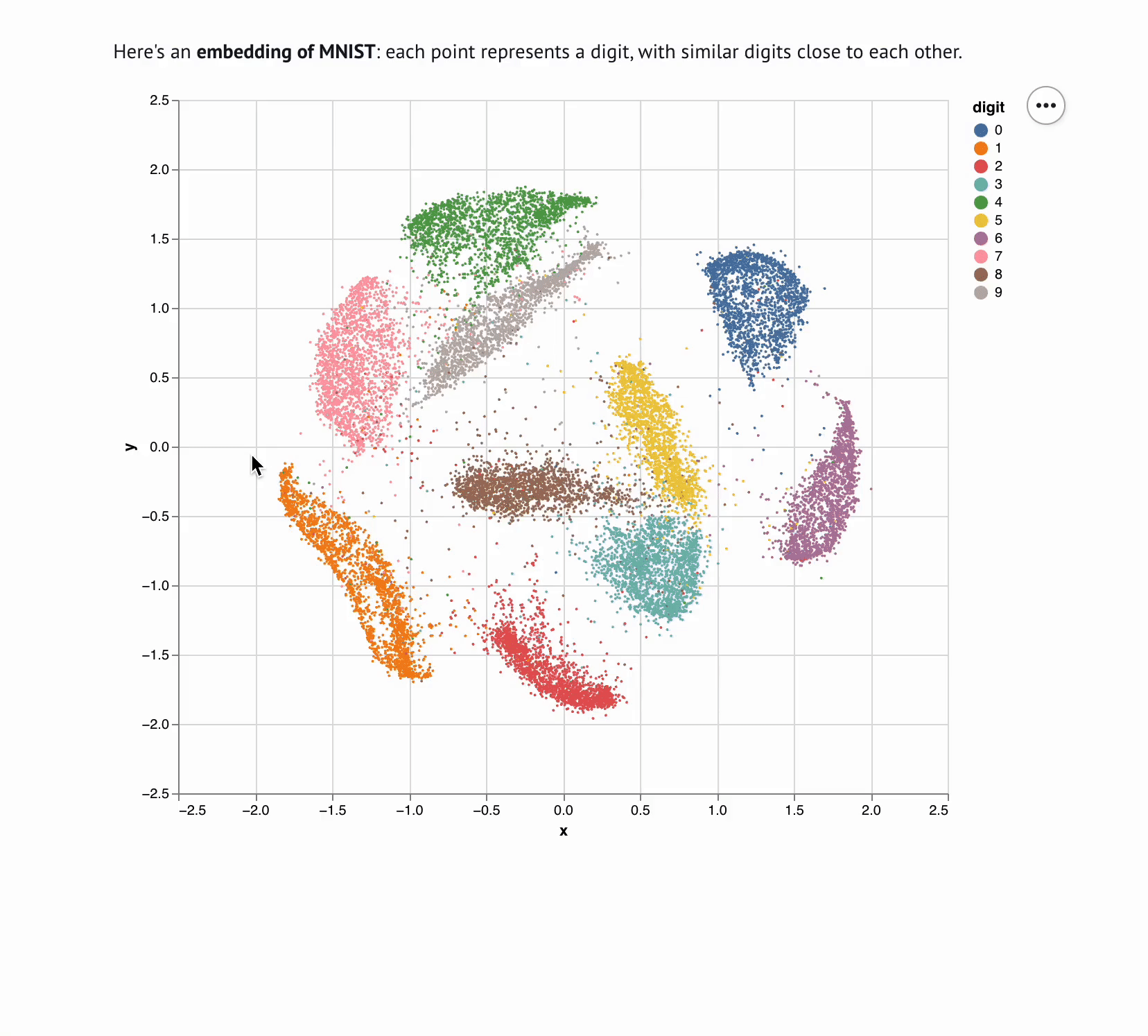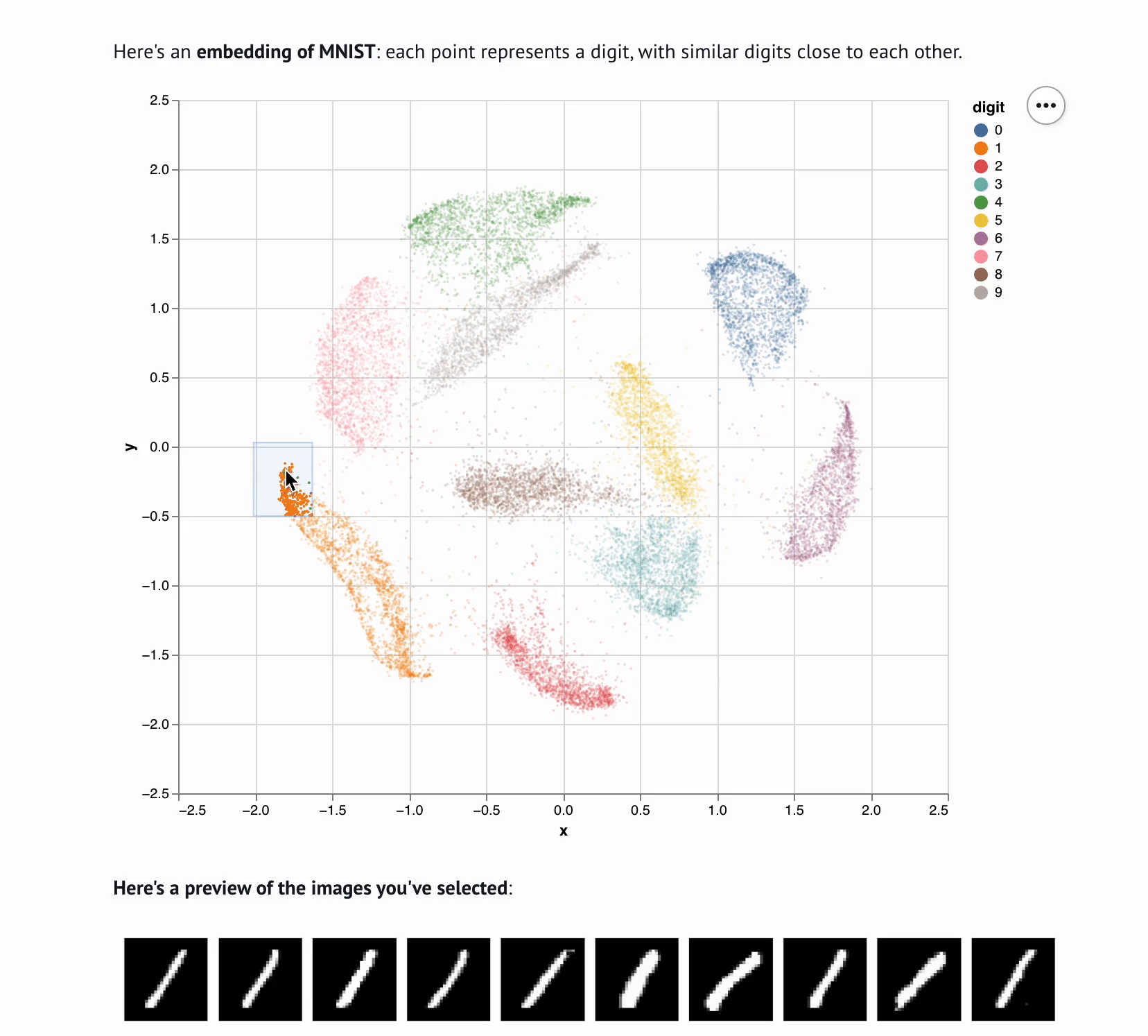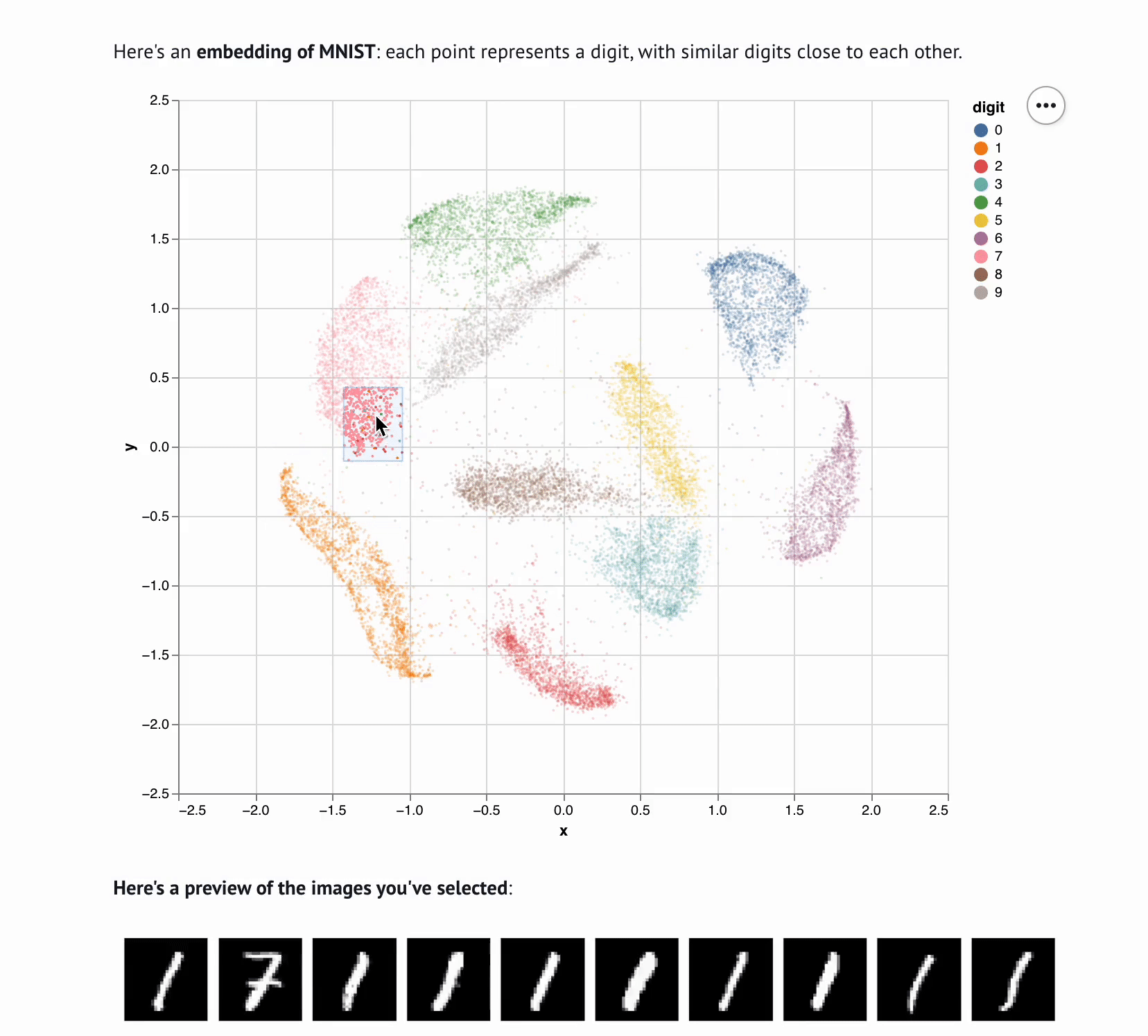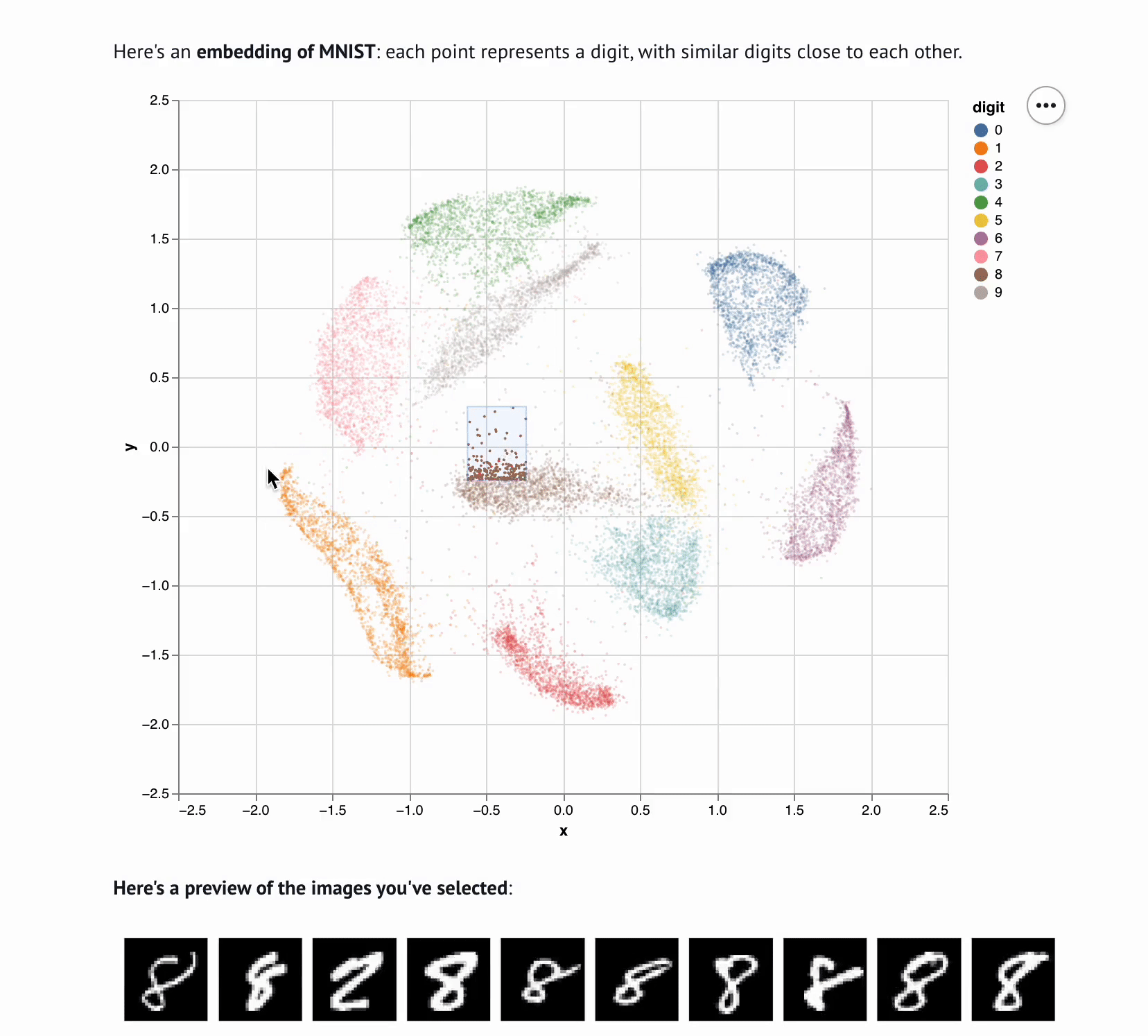
+**This template lets you visualize and interactively explore high dimensional
+data.** The starter code uses PCA to embed and plot numerical digits, seeing
+how they cluster together — when you select points in the plot, the notebook
+shows you the underlying images!
+
+To use this notebook on your own data, just replace the implementations
+of the following four functions:
+
+* `load_data`
+* `embed_data`
+* `scatter_data`
+* `show_selection`
+
+ +
+## Running this notebook
+
+Open this notebook in [our online
+playground](https://marimo.app/github.com/marimo-team/examples/blob/main/explore_high_dimensional_data/explore_high_dimensional_data.py)
+or run it locally.
+
+### Running locally
+
+The requirements of each notebook are serialized in them as a top-level
+comment. Here are the steps to run the notebook:
+
+1. [Install `uv`](https://github.com/astral-sh/uv/?tab=readme-ov-file#installation)
+2. Open an example with `uvx marimo edit --sandbox `
+
+> [!TIP]
+> The [`--sandbox`
+> flag](https://docs.marimo.io/guides/package_reproducibility/) opens the
+> notebook in an isolated virtual environment, automatically installing the
+> notebook's dependencies 📦
+
+You can also open notebooks without `uv`, in which case you'll need to
+manually [install marimo](https://docs.marimo.io/getting_started/index.html#installation)
+first. Then run `marimo edit `; however, you'll also need to
+install the requirements yourself.
diff --git a/explore_high_dimensional_data/explore_high_dimensional_data.py b/explore_high_dimensional_data/explore_high_dimensional_data.py
new file mode 100644
index 0000000..15d8430
--- /dev/null
+++ b/explore_high_dimensional_data/explore_high_dimensional_data.py
@@ -0,0 +1,226 @@
+# /// script
+# requires-python = ">=3.12"
+# dependencies = [
+# "altair==5.5.0",
+# "marimo",
+# "matplotlib==3.10.0",
+# "pandas==2.2.3",
+# "polars==1.20.0",
+# "scikit-learn==1.6.1",
+# ]
+# ///
+
+import marimo
+
+__generated_with = "0.10.16"
+app = marimo.App(width="columns")
+
+
+@app.cell(column=0, hide_code=True)
+def _(mo):
+ mo.md(
+ """
+ **This template lets you visualize and interactively explore high dimensional data.** The starter code uses PCA to embed and plot numerical
+ digits, seeing how they cluster together — when you select points in the plot, the notebook shows you the underlying images.
+
+ The left-hand column implements the core logic; the right-hand column executes the dimensionality reduction and shows the outputs.
+
+ **To customize this template to your own data, just implement the functions in
+ this column.**
+ """
+ )
+ return
+
+
+@app.cell
+def _():
+ def load_data():
+ """
+ Return a tuple of your data:
+
+ * The dataset, with each row a different item in the dataset
+ * A label for each data point.
+
+ If your data doesn't have labels, just return a list of all ones
+ with the same length as the number of items in your dataset.
+ """
+ import sklearn.datasets
+
+ data, labels = sklearn.datasets.load_digits(return_X_y=True)
+ return data, labels
+
+ return (load_data,)
+
+
+@app.cell
+def _():
+ def embed_data(data):
+ """
+ Embed the data into two dimensions. The default implementation
+ uses PCA, but you can also use UMAP, tSNE, or any other dimensionality
+ reduction algorithm you like.
+
+ The starter implementation here uses PCA, and assumes the data is a NumPy
+ array.
+ """
+ import sklearn
+
+ return sklearn.decomposition.PCA(n_components=2, whiten=True).fit_transform(
+ data
+ )
+
+ return (embed_data,)
+
+
+@app.cell
+def _(pl):
+ def scatter_data(df: pl.DataFrame) -> alt.Chart:
+ """
+ Visualize the embedded data using an Altair scatterplot.
+
+ - df is a Polars dataframe with the following columns:
+ * x: the first coordinate of the embedding
+ * y: the second coordinate of the embedding
+ * label: a label identifying each item, for coloring
+
+ Modify the starter implementation to suit your needs, but make sure
+ to return an altair chart.
+ """
+ import altair as alt
+
+ return (
+ alt.Chart(df)
+ .mark_circle()
+ .encode(
+ x=alt.X("x:Q").scale(domain=(-2.5, 2.5)),
+ y=alt.Y("y:Q").scale(domain=(-2.5, 2.5)),
+ color=alt.Color("label:N"),
+ )
+ .properties(width=500, height=500)
+ )
+
+ return (scatter_data,)
+
+
+@app.cell
+def _():
+ def show_selection(data, rows, max_rows=10):
+ """
+ Visualize selected rows of the data.
+
+ - `data` is the data returned from `load_data`
+ - `rows` is a list or array of row indices
+ - `max_rows` is the maximum number of rows to display
+ """
+ import matplotlib.pyplot as plt
+
+ # show 10 images: either the first 10 from the selection, or the first ten
+ # selected in the table
+ rows = rows[:max_rows]
+ images = data.reshape((-1, 8, 8))[rows]
+ fig, axes = plt.subplots(1, len(rows))
+ fig.set_size_inches(12.5, 1.5)
+ if len(rows) > 1:
+ for im, ax in zip(images, axes.flat):
+ ax.imshow(im, cmap="gray")
+ ax.set_yticks([])
+ ax.set_xticks([])
+ else:
+ axes.imshow(images[0], cmap="gray")
+ axes.set_yticks([])
+ axes.set_xticks([])
+ plt.tight_layout()
+ return fig
+
+ return (show_selection,)
+
+
+@app.cell(column=1, hide_code=True)
+def _(mo):
+ mo.md("""# Explore high dimensional data""")
+ return
+
+
+@app.cell(hide_code=True)
+def _(mo):
+ mo.md(
+ """
+ Here's an **embedding** of your data, with similar points close to each other.
+
+ This notebook will automatically drill down into points you **select with
+ your mouse**; try it!
+ """
+ )
+ return
+
+
+@app.cell
+def _(load_data):
+ data, labels = load_data()
+ return data, labels
+
+
+@app.cell
+def _():
+ import polars as pl
+
+ return (pl,)
+
+
+@app.cell
+def _(data, embed_data, labels, pl):
+ X_embedded = embed_data(data)
+
+ embedding = pl.DataFrame(
+ {
+ "x": X_embedded[:, 0],
+ "y": X_embedded[:, 1],
+ "label": labels,
+ "index": list(range(X_embedded.shape[0])),
+ }
+ )
+ return X_embedded, embedding
+
+
+@app.cell
+def _(embedding, mo, scatter_data):
+ chart = mo.ui.altair_chart(scatter_data(embedding))
+ chart
+ return (chart,)
+
+
+@app.cell
+def _(chart, mo):
+ table = mo.ui.table(chart.value)
+ return (table,)
+
+
+@app.cell
+def _(chart, data, mo, show_selection, table):
+ mo.stop(not len(chart.value))
+
+ selected_rows = show_selection(data, list(chart.value["index"]))
+
+ mo.md(
+ f"""
+ **Here's a preview of the items you've selected**:
+
+ {mo.as_html(selected_rows)}
+
+ Here's all the data you've selected.
+
+ {table}
+ """
+ )
+ return (selected_rows,)
+
+
+@app.cell
+def _():
+ import marimo as mo
+
+ return (mo,)
+
+
+if __name__ == "__main__":
+ app.run()
diff --git a/nlp_span_comparison/README.md b/nlp_span_comparison/README.md
index 25e14b6..75a2248 100644
--- a/nlp_span_comparison/README.md
+++ b/nlp_span_comparison/README.md
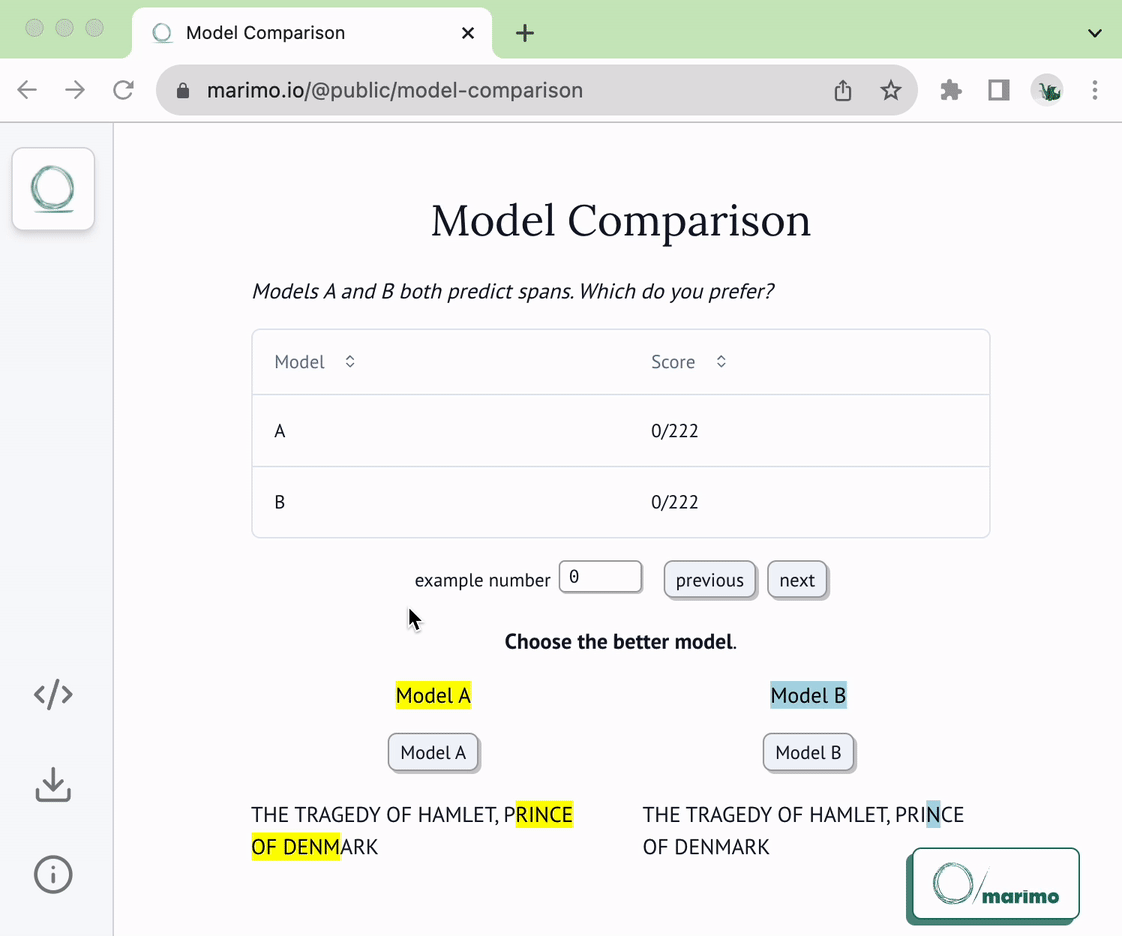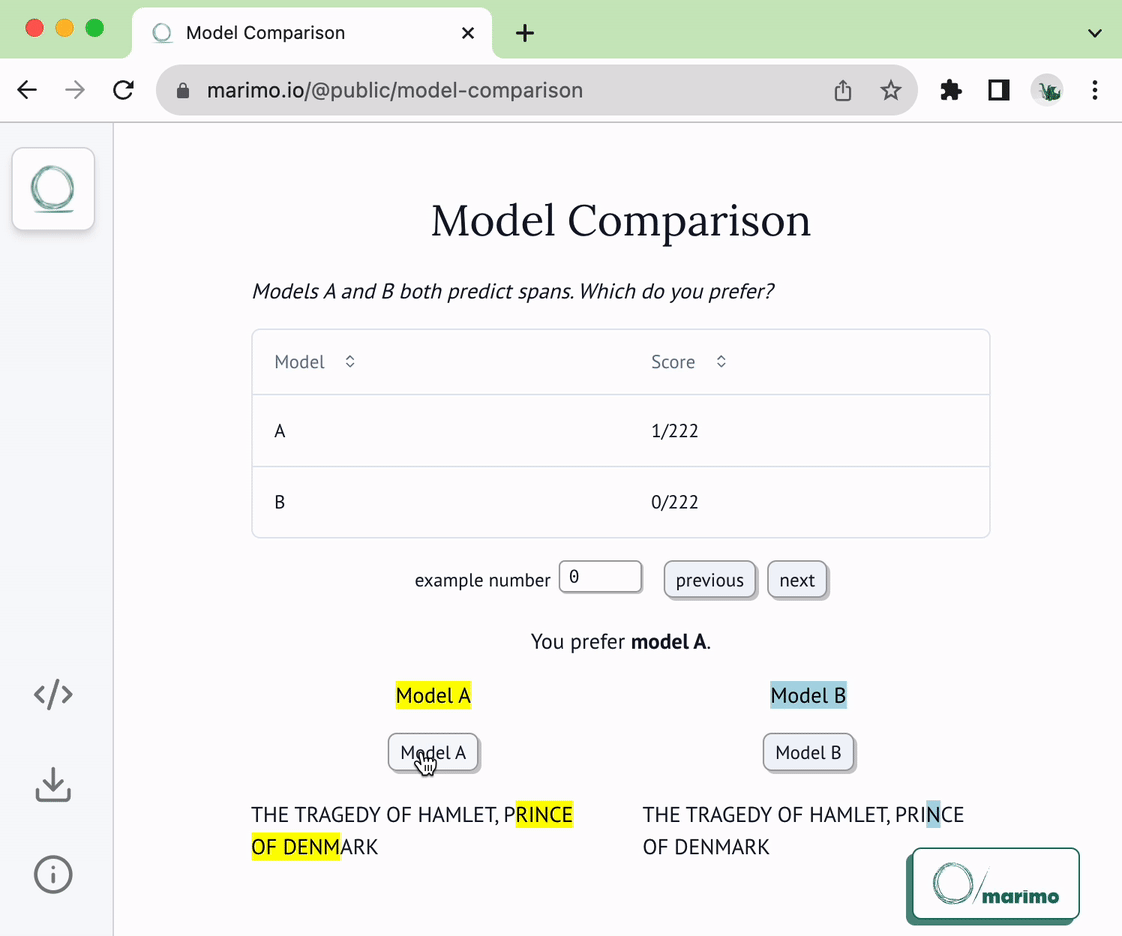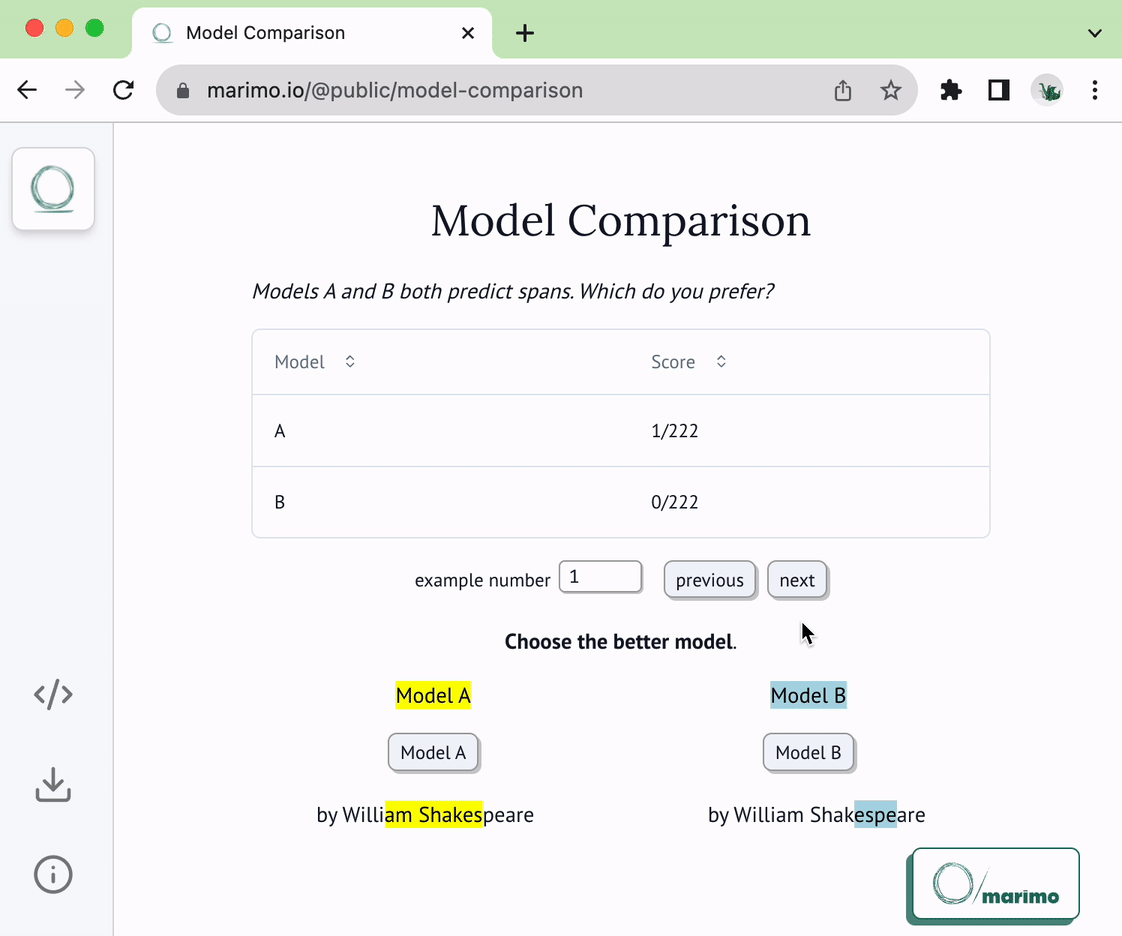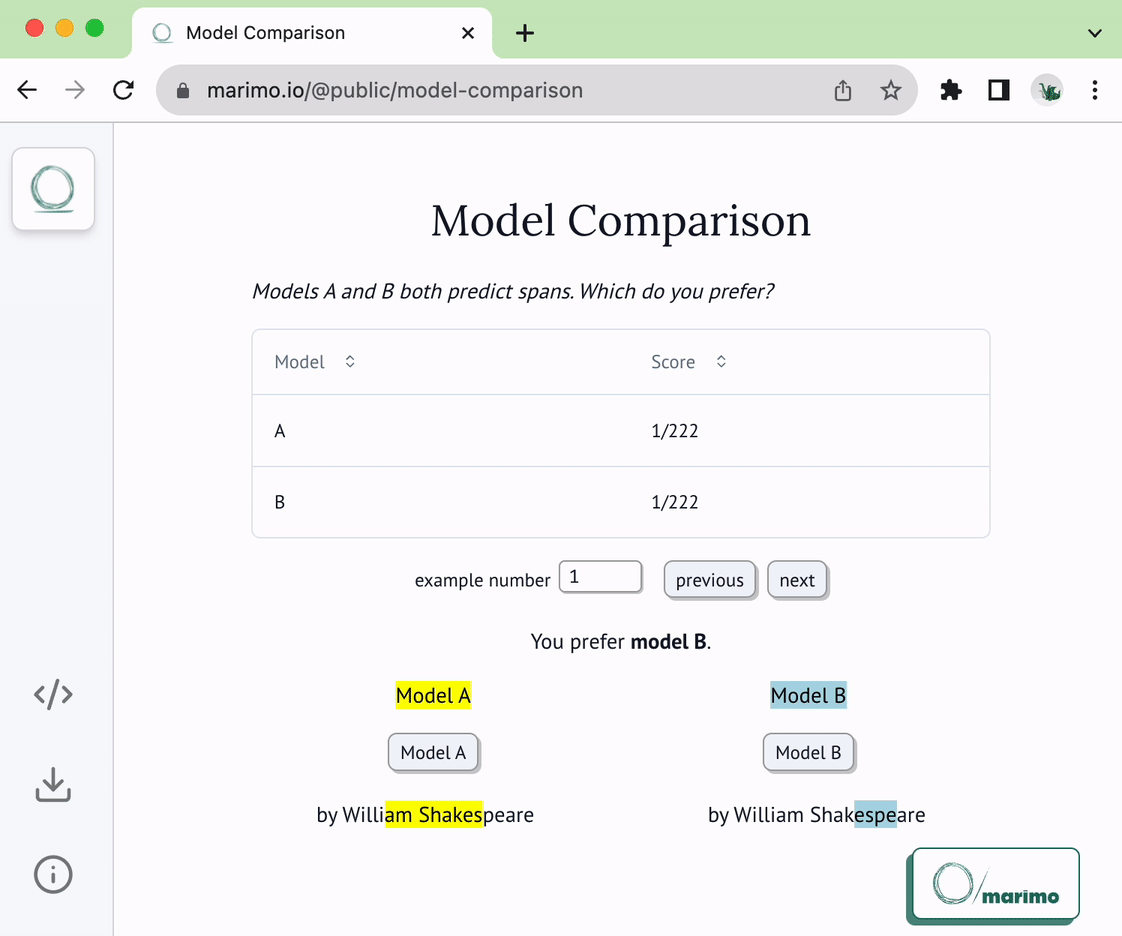
@@ -22,6 +22,8 @@ of the following two functions:
* `load_choices`
* `save_choices`
+
+
+## Running this notebook
+
+Open this notebook in [our online
+playground](https://marimo.app/github.com/marimo-team/examples/blob/main/explore_high_dimensional_data/explore_high_dimensional_data.py)
+or run it locally.
+
+### Running locally
+
+The requirements of each notebook are serialized in them as a top-level
+comment. Here are the steps to run the notebook:
+
+1. [Install `uv`](https://github.com/astral-sh/uv/?tab=readme-ov-file#installation)
+2. Open an example with `uvx marimo edit --sandbox `
+
+> [!TIP]
+> The [`--sandbox`
+> flag](https://docs.marimo.io/guides/package_reproducibility/) opens the
+> notebook in an isolated virtual environment, automatically installing the
+> notebook's dependencies 📦
+
+You can also open notebooks without `uv`, in which case you'll need to
+manually [install marimo](https://docs.marimo.io/getting_started/index.html#installation)
+first. Then run `marimo edit `; however, you'll also need to
+install the requirements yourself.
diff --git a/explore_high_dimensional_data/explore_high_dimensional_data.py b/explore_high_dimensional_data/explore_high_dimensional_data.py
new file mode 100644
index 0000000..15d8430
--- /dev/null
+++ b/explore_high_dimensional_data/explore_high_dimensional_data.py
@@ -0,0 +1,226 @@
+# /// script
+# requires-python = ">=3.12"
+# dependencies = [
+# "altair==5.5.0",
+# "marimo",
+# "matplotlib==3.10.0",
+# "pandas==2.2.3",
+# "polars==1.20.0",
+# "scikit-learn==1.6.1",
+# ]
+# ///
+
+import marimo
+
+__generated_with = "0.10.16"
+app = marimo.App(width="columns")
+
+
+@app.cell(column=0, hide_code=True)
+def _(mo):
+ mo.md(
+ """
+ **This template lets you visualize and interactively explore high dimensional data.** The starter code uses PCA to embed and plot numerical
+ digits, seeing how they cluster together — when you select points in the plot, the notebook shows you the underlying images.
+
+ The left-hand column implements the core logic; the right-hand column executes the dimensionality reduction and shows the outputs.
+
+ **To customize this template to your own data, just implement the functions in
+ this column.**
+ """
+ )
+ return
+
+
+@app.cell
+def _():
+ def load_data():
+ """
+ Return a tuple of your data:
+
+ * The dataset, with each row a different item in the dataset
+ * A label for each data point.
+
+ If your data doesn't have labels, just return a list of all ones
+ with the same length as the number of items in your dataset.
+ """
+ import sklearn.datasets
+
+ data, labels = sklearn.datasets.load_digits(return_X_y=True)
+ return data, labels
+
+ return (load_data,)
+
+
+@app.cell
+def _():
+ def embed_data(data):
+ """
+ Embed the data into two dimensions. The default implementation
+ uses PCA, but you can also use UMAP, tSNE, or any other dimensionality
+ reduction algorithm you like.
+
+ The starter implementation here uses PCA, and assumes the data is a NumPy
+ array.
+ """
+ import sklearn
+
+ return sklearn.decomposition.PCA(n_components=2, whiten=True).fit_transform(
+ data
+ )
+
+ return (embed_data,)
+
+
+@app.cell
+def _(pl):
+ def scatter_data(df: pl.DataFrame) -> alt.Chart:
+ """
+ Visualize the embedded data using an Altair scatterplot.
+
+ - df is a Polars dataframe with the following columns:
+ * x: the first coordinate of the embedding
+ * y: the second coordinate of the embedding
+ * label: a label identifying each item, for coloring
+
+ Modify the starter implementation to suit your needs, but make sure
+ to return an altair chart.
+ """
+ import altair as alt
+
+ return (
+ alt.Chart(df)
+ .mark_circle()
+ .encode(
+ x=alt.X("x:Q").scale(domain=(-2.5, 2.5)),
+ y=alt.Y("y:Q").scale(domain=(-2.5, 2.5)),
+ color=alt.Color("label:N"),
+ )
+ .properties(width=500, height=500)
+ )
+
+ return (scatter_data,)
+
+
+@app.cell
+def _():
+ def show_selection(data, rows, max_rows=10):
+ """
+ Visualize selected rows of the data.
+
+ - `data` is the data returned from `load_data`
+ - `rows` is a list or array of row indices
+ - `max_rows` is the maximum number of rows to display
+ """
+ import matplotlib.pyplot as plt
+
+ # show 10 images: either the first 10 from the selection, or the first ten
+ # selected in the table
+ rows = rows[:max_rows]
+ images = data.reshape((-1, 8, 8))[rows]
+ fig, axes = plt.subplots(1, len(rows))
+ fig.set_size_inches(12.5, 1.5)
+ if len(rows) > 1:
+ for im, ax in zip(images, axes.flat):
+ ax.imshow(im, cmap="gray")
+ ax.set_yticks([])
+ ax.set_xticks([])
+ else:
+ axes.imshow(images[0], cmap="gray")
+ axes.set_yticks([])
+ axes.set_xticks([])
+ plt.tight_layout()
+ return fig
+
+ return (show_selection,)
+
+
+@app.cell(column=1, hide_code=True)
+def _(mo):
+ mo.md("""# Explore high dimensional data""")
+ return
+
+
+@app.cell(hide_code=True)
+def _(mo):
+ mo.md(
+ """
+ Here's an **embedding** of your data, with similar points close to each other.
+
+ This notebook will automatically drill down into points you **select with
+ your mouse**; try it!
+ """
+ )
+ return
+
+
+@app.cell
+def _(load_data):
+ data, labels = load_data()
+ return data, labels
+
+
+@app.cell
+def _():
+ import polars as pl
+
+ return (pl,)
+
+
+@app.cell
+def _(data, embed_data, labels, pl):
+ X_embedded = embed_data(data)
+
+ embedding = pl.DataFrame(
+ {
+ "x": X_embedded[:, 0],
+ "y": X_embedded[:, 1],
+ "label": labels,
+ "index": list(range(X_embedded.shape[0])),
+ }
+ )
+ return X_embedded, embedding
+
+
+@app.cell
+def _(embedding, mo, scatter_data):
+ chart = mo.ui.altair_chart(scatter_data(embedding))
+ chart
+ return (chart,)
+
+
+@app.cell
+def _(chart, mo):
+ table = mo.ui.table(chart.value)
+ return (table,)
+
+
+@app.cell
+def _(chart, data, mo, show_selection, table):
+ mo.stop(not len(chart.value))
+
+ selected_rows = show_selection(data, list(chart.value["index"]))
+
+ mo.md(
+ f"""
+ **Here's a preview of the items you've selected**:
+
+ {mo.as_html(selected_rows)}
+
+ Here's all the data you've selected.
+
+ {table}
+ """
+ )
+ return (selected_rows,)
+
+
+@app.cell
+def _():
+ import marimo as mo
+
+ return (mo,)
+
+
+if __name__ == "__main__":
+ app.run()
diff --git a/nlp_span_comparison/README.md b/nlp_span_comparison/README.md
index 25e14b6..75a2248 100644
--- a/nlp_span_comparison/README.md
+++ b/nlp_span_comparison/README.md
@@ -22,6 +22,8 @@ of the following two functions:
* `load_choices`
* `save_choices`
+ +
## Running this notebook
Open this notebook in [our online
+
## Running this notebook
Open this notebook in [our online
 +
## Running this notebook
Open this notebook in [our online
+
## Running this notebook
Open this notebook in [our online
 +
+## Running this notebook
+
+Open this notebook in [our online
+playground](https://marimo.app/github.com/marimo-team/examples/blob/main/explore_high_dimensional_data/explore_high_dimensional_data.py)
+or run it locally.
+
+### Running locally
+
+The requirements of each notebook are serialized in them as a top-level
+comment. Here are the steps to run the notebook:
+
+1. [Install `uv`](https://github.com/astral-sh/uv/?tab=readme-ov-file#installation)
+2. Open an example with `uvx marimo edit --sandbox
+
+## Running this notebook
+
+Open this notebook in [our online
+playground](https://marimo.app/github.com/marimo-team/examples/blob/main/explore_high_dimensional_data/explore_high_dimensional_data.py)
+or run it locally.
+
+### Running locally
+
+The requirements of each notebook are serialized in them as a top-level
+comment. Here are the steps to run the notebook:
+
+1. [Install `uv`](https://github.com/astral-sh/uv/?tab=readme-ov-file#installation)
+2. Open an example with `uvx marimo edit --sandbox  +
## Running this notebook
Open this notebook in [our online
+
## Running this notebook
Open this notebook in [our online