You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
It then runs the jupyter notebook without problems.
So I follow the preprocessing code from nipype_tutorial
You can see my code below:
from os.path import join as opj
import os
import json
from nipype.interfaces.fsl import (BET, ExtractROI, FAST, FLIRT, ImageMaths,
MCFLIRT, SliceTimer, Threshold)
from nipype.interfaces.spm import Smooth
from nipype.interfaces.utility import IdentityInterface
from nipype.interfaces.io import SelectFiles, DataSink
from nipype.algorithms.rapidart import ArtifactDetect
from nipype import Workflow, Node
experiment_dir = '/home/neuro/nipype_tutorial/output'
output_dir = 'datasink'
working_dir = 'workingdir'
# list of subject identifiers
subject_list = ['01', '02', '03', '04', '05', '06', '07', '08', '09', '10', '11', '12', '13',
'14', '15', '16', '17', '18', '19', '20', '21', '22', '23','24','25','26']
# list of session identifiers
task_list = ['motorseq']
# Smoothing widths to apply
fwhm = [4, 8]
# TR of functional images
with open('/home/neuro/nipype_tutorial/data/task-motorseq_bold.json', 'rt') as fp:
task_info = json.load(fp)
TR = task_info['RepetitionTime']
# Isometric resample of functional images to voxel size (in mm)
iso_size = 4
# ExtractROI - skip dummy scans
extract = Node(ExtractROI(t_min=4, t_size=-1, output_type='NIFTI'),
name="extract")
# MCFLIRT - motion correction
mcflirt = Node(MCFLIRT(mean_vol=True,
save_plots=True,
output_type='NIFTI'),
name="mcflirt")
# SliceTimer - correct for slice wise acquisition
slicetimer = Node(SliceTimer(index_dir=False,
interleaved=True,
output_type='NIFTI',
time_repetition=TR),
name="slicetimer")
# Smooth - image smoothing
smooth = Node(Smooth(), name="smooth")
smooth.iterables = ("fwhm", fwhm)
# Artifact Detection - determines outliers in functional images
art = Node(ArtifactDetect(norm_threshold=2,
zintensity_threshold=3,
mask_type='spm_global',
parameter_source='FSL',
use_differences=[True, False],
plot_type='svg'),
name="art")
# BET - Skullstrip anatomical Image
bet_anat = Node(BET(frac=0.5,
robust=True,
output_type='NIFTI_GZ'),
name="bet_anat")
# FAST - Image Segmentation
segmentation = Node(FAST(output_type='NIFTI_GZ'),
name="segmentation")
# Select WM segmentation file from segmentation output
def get_wm(files):
return files[-1]
# Threshold - Threshold WM probability image
threshold = Node(Threshold(thresh=0.5,
args='-bin',
output_type='NIFTI_GZ'),
name="threshold")
# FLIRT - pre-alignment of functional images to anatomical images
coreg_pre = Node(FLIRT(dof=6, output_type='NIFTI_GZ'),
name="coreg_pre")
# FLIRT - coregistration of functional images to anatomical images with BBR
coreg_bbr = Node(FLIRT(dof=6,
cost='bbr',
schedule=opj(os.getenv('FSLDIR'),
'etc/flirtsch/bbr.sch'),
output_type='NIFTI_GZ'),
name="coreg_bbr")
# Apply coregistration warp to functional images
applywarp = Node(FLIRT(interp='spline',
apply_isoxfm=iso_size,
output_type='NIFTI'),
name="applywarp")
# Apply coregistration warp to mean file
applywarp_mean = Node(FLIRT(interp='spline',
apply_isoxfm=iso_size,
output_type='NIFTI_GZ'),
name="applywarp_mean")
# Create a coregistration workflow
coregwf = Workflow(name='coregwf')
coregwf.base_dir = opj(experiment_dir, working_dir)
# Connect all components of the coregistration workflow
coregwf.connect([(bet_anat, segmentation, [('out_file', 'in_files')]),
(segmentation, threshold, [(('partial_volume_files', get_wm),
'in_file')]),
(bet_anat, coreg_pre, [('out_file', 'reference')]),
(threshold, coreg_bbr, [('out_file', 'wm_seg')]),
(coreg_pre, coreg_bbr, [('out_matrix_file', 'in_matrix_file')]),
(coreg_bbr, applywarp, [('out_matrix_file', 'in_matrix_file')]),
(bet_anat, applywarp, [('out_file', 'reference')]),
(coreg_bbr, applywarp_mean, [('out_matrix_file', 'in_matrix_file')]),
(bet_anat, applywarp_mean, [('out_file', 'reference')]),
])
runs = ['01', '02', '03', '04', '05', '06', '07', '08']
sessions = ['01', '02', '03', '04']
# Infosource - a function free node to iterate over the list of subject names
infosource = Node(IdentityInterface(fields=['subject_id', 'task_name', 'run_list','session_list']),
name="infosource")
infosource.iterables = [('subject_id', subject_list),
('task_name', task_list),
('run_list', runs),
('session_list', sessions)
]
#### sub-{subject_id}_ses-test_task-{task_name}_bold.nii.gz
### sub-{subject_id}_ses-{session_list}_task-{task_name}_run-{run_list}_bold.nii.gz
# SelectFiles - to grab the data (alternativ to DataGrabber)
anat_file = opj('sub-{subject_id}', 'anat', 'sub-{subject_id}_ses-{session_list}_T1w.nii.gz')
func_file = opj('sub-{subject_id}', 'func',
'sub-{subject_id}_ses-{session_list}_task-{task_name}_run-{run_list}_bold.nii.gz')
templates = {'anat': anat_file,
'func': func_file}
selectfiles = Node(SelectFiles(templates,
base_directory='/home/neuro/nipype_tutorial/data'),
name="selectfiles")
# Datasink - creates output folder for important outputs
datasink = Node(DataSink(base_directory=experiment_dir,
container=output_dir),
name="datasink")
## Use the following DataSink output substitutions
substitutions = [('_subject_id_', 'sub-'),
('_task_name_', '/task-'),
('_fwhm_', 'fwhm-'),
('_roi', ''),
('_mcf', ''),
('_st', ''),
('_flirt', ''),
('.nii_mean_reg', '_mean'),
('.nii.par', '.par'),
]
subjFolders = [('fwhm-%s/' % f, 'fwhm-%s_' % f) for f in fwhm]
substitutions.extend(subjFolders)
datasink.inputs.substitutions = substitutions
# Create a preprocessing workflow
preproc = Workflow(name='preproc')
preproc.base_dir = opj(experiment_dir, working_dir)
# Connect all components of the preprocessing workflow
preproc.connect([(infosource, selectfiles, [('subject_id', 'subject_id'),
('task_name', 'task_name')]),
(selectfiles, extract, [('func', 'in_file')]),
(extract, mcflirt, [('roi_file', 'in_file')]),
(mcflirt, slicetimer, [('out_file', 'in_file')]),
(selectfiles, coregwf, [('anat', 'bet_anat.in_file'),
('anat', 'coreg_bbr.reference')]),
(mcflirt, coregwf, [('mean_img', 'coreg_pre.in_file'),
('mean_img', 'coreg_bbr.in_file'),
('mean_img', 'applywarp_mean.in_file')]),
(slicetimer, coregwf, [('slice_time_corrected_file', 'applywarp.in_file')]),
(coregwf, smooth, [('applywarp.out_file', 'in_files')]),
(mcflirt, datasink, [('par_file', 'preproc.@par')]),
(smooth, datasink, [('smoothed_files', 'preproc.@smooth')]),
(coregwf, datasink, [('applywarp_mean.out_file', 'preproc.@mean')]),
(coregwf, art, [('applywarp.out_file', 'realigned_files')]),
(mcflirt, art, [('par_file', 'realignment_parameters')]),
(coregwf, datasink, [('coreg_bbr.out_matrix_file', 'preproc.@mat_file'),
('bet_anat.out_file', 'preproc.@brain')]),
(art, datasink, [('outlier_files', 'preproc.@outlier_files'),
('plot_files', 'preproc.@plot_files')]),
])
# Create preproc output graph
preproc.write_graph(graph2use='colored', format='png', simple_form=True)
# Visualize the graph
from IPython.display import Image
Image(filename=opj(preproc.base_dir, 'preproc', 'graph.png'))
# Visualize the detailed graph
preproc.write_graph(graph2use='flat', format='png', simple_form=True)
Image(filename=opj(preproc.base_dir, 'preproc', 'graph_detailed.png'))
preproc.run('MultiProc', plugin_args={'n_procs': 8})
Then a big error comes and I don't know how to solve:
230214-23:22:00,267 nipype.workflow INFO:
Workflow preproc settings: ['check', 'execution', 'logging', 'monitoring']
230214-23:22:15,779 nipype.workflow INFO:
Running in parallel.
230214-23:22:16,108 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 832 jobs ready. Free memory (GB): 6.99/6.99, Free processors: 8/8.
230214-23:22:16,243 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_08_session_list_02_subject_id_26_task_name_motorseq/selectfiles".230214-23:22:16,247 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_07_session_list_03_subject_id_26_task_name_motorseq/selectfiles".230214-23:22:16,248 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_07_session_list_02_subject_id_26_task_name_motorseq/selectfiles".
230214-23:22:16,245 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_08_session_list_01_subject_id_26_task_name_motorseq/selectfiles".230214-23:22:16,247 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_07_session_list_04_subject_id_26_task_name_motorseq/selectfiles".
230214-23:22:16,254 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")230214-23:22:16,258 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")230214-23:22:16,261 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
230214-23:22:16,267 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
230214-23:22:16,269 nipype.workflow WARNING:
Storing result file without outputs230214-23:22:16,273 nipype.workflow WARNING:
Storing result file without outputs
230214-23:22:16,262 nipype.workflow INFO:
[Node] Running "selectfiles" ("nipype.interfaces.io.SelectFiles")
230214-23:22:16,277 nipype.workflow WARNING:
Storing result file without outputs230214-23:22:16,241 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_08_session_list_03_subject_id_26_task_name_motorseq/selectfiles".230214-23:22:16,239 nipype.workflow INFO:
[Node] Setting-up "preproc.selectfiles" in "/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_08_session_list_04_subject_id_26_task_name_motorseq/selectfiles".
230214-23:22:16,280 nipype.workflow WARNING:
Storing result file without outputs
230214-23:22:16,277 nipype.workflow WARNING:
[Node] Error on "preproc.selectfiles" (/home/neuro/nipype_tutorial/output/workingdir/preproc/_run_list_07_session_list_02_subject_id_26_task_name_motorseq/selectfiles)230214-23:22:16,288 nipype.workflow WARNING:
Storing result file without outputs
There is more, but these are the first errors the appear. Seems like something about the folders or path, I tired changing base directory, but it did not solve.
Can anyone #help?
The text was updated successfully, but these errors were encountered:
Hi, I have this bids file:
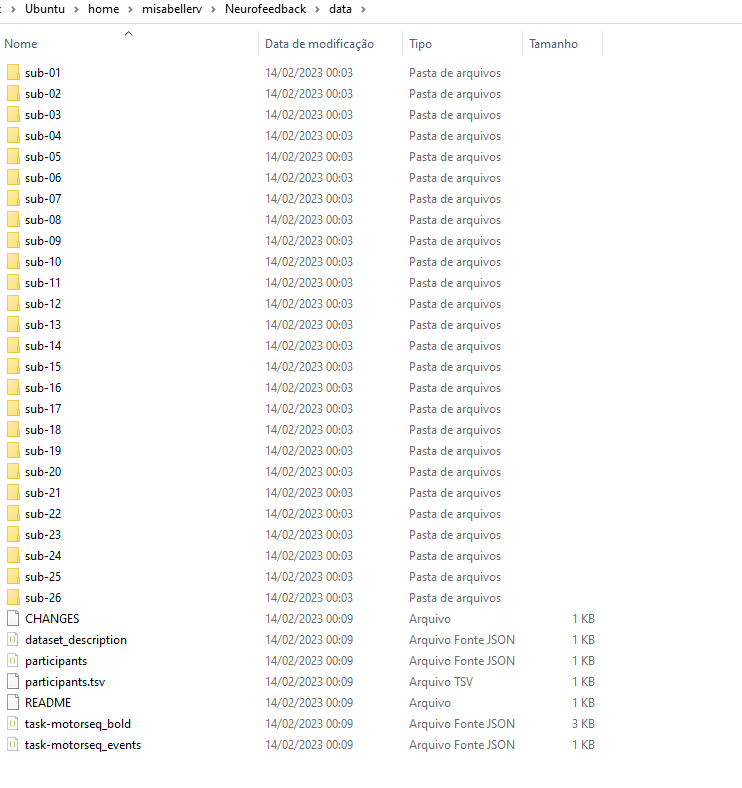
Each sub-* has 4 sessions:
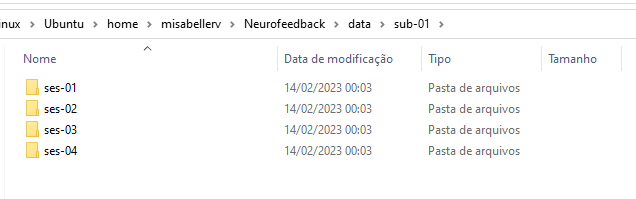
composed of anat, func, and fmap folders.
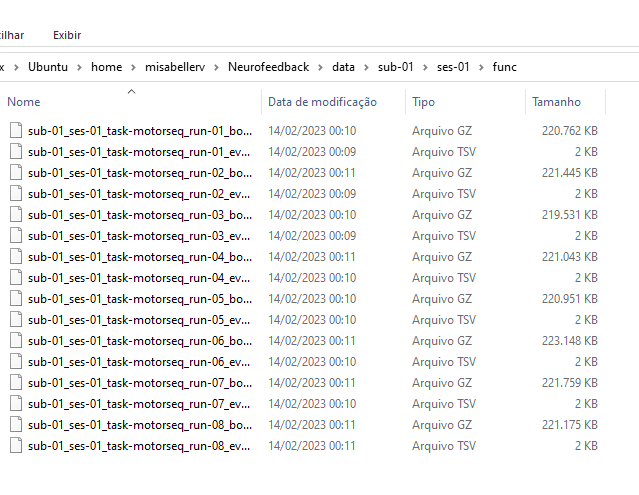
Each func folder has 8 runs:
and I'm trying to run preprocessing workflow from nipype_tutorial but it fails and I can't fix/find the reason.
First i open the jupyter notebook for my local data using:
docker run -it -v /home/misabellerv/Neurofeedback/:/home/neuro/nipype_tutorial -v /home/misabellerv/Neurofeedback/data/:/data -v /home/misabellerv/Neurofeedback/output/:/output -p 8888:8888 miykael/nipype_tutorial jupyter notebookIt then runs the jupyter notebook without problems.
So I follow the preprocessing code from nipype_tutorial
You can see my code below:
Then a big error comes and I don't know how to solve:
There is more, but these are the first errors the appear. Seems like something about the folders or path, I tired changing base directory, but it did not solve.
Can anyone #help?
The text was updated successfully, but these errors were encountered: