-
Notifications
You must be signed in to change notification settings - Fork 0
/
README.Rmd
142 lines (96 loc) · 6.01 KB
/
README.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
---
output: github_document
---
<!-- README.md is generated from README.Rmd. Please edit that file -->
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
# nonmem2rx
<!-- badges: start -->
[](https://github.com/nlmixr2/nonmem2rx/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/nlmixr2/nonmem2rx?branch=main)
[](https://cran.r-project.org/package=nonmem2rx)
[](https://cran.r-project.org/package=nonmem2rx)
[](https://cran.r-project.org/package=nonmem2rx)
[](https://www.codefactor.io/repository/github/nlmixr2/nonmem2rx)

<!-- badges: end -->
The goal of nonmem2rx is to convert a NONMEM control stream to a
`rxode2` (or even a `nlmixr2` fit) for easy clinical trial simulation
in R.
## Installation
You can install the development version of nonmem2rx from
[GitHub](https://github.com/) with the r-universe:
``` r
install.packages('nonmem2rx', repos = c('https://nlmixr2.r-universe.dev', 'https://cloud.r-project.org'))
```
You can also get the CRAN version by:
``` r
install.packages('nonmem2rx')
```
## What you can do with `nonmem2rx`/`babelmixr2`
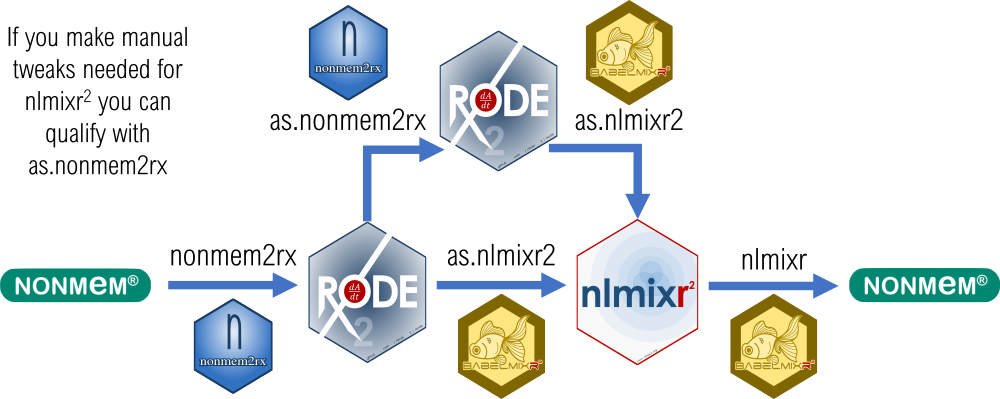
You can do many useful tasks directly converting between nlmixr2 and NONMEM models; you can:
- [Convert a NONMEM model to a rxode2 model](https://nlmixr2.github.io/nonmem2rx/articles/import-nonmem.html)
- [Do development in nlmixr2 and then run NONMEM from a nlmixr2 model](https://nlmixr2.github.io/babelmixr2/articles/running-nonmem.html) for reviewers who want to know about NONMEM results.
- In both conversions, [automatically make sure the model is translated correctly ](https://nlmixr2.github.io/nonmem2rx/articles/rxode2-validate.html) (for [babelmixr2](https://nlmixr2.github.io/babelmixr2/articles/running-nonmem.html#optional-step-2-recover-a-failed-nonmem-run))
Then with nlmixr2 fit models and nonmem2rx models coming from both conversions, you can:
- [Perform simulations of new
dosing](https://nlmixr2.github.io/nonmem2rx/articles/simulate-new-dosing.html) from the NONMEM model or even [simulate using the uncertainty in your model to simulate new scenarios](https://nlmixr2.github.io/nonmem2rx/articles/simulate-uncertainty.html)
- [Modify the model to calculate derived parameters](https://nlmixr2.github.io/nonmem2rx/articles/simulate-extra-items.html) (like AUC). These parameters slow down NONMEM’s optimization, but can help in your simulation scenario.
- [Simulating with Covariates/Input PK
parameters](https://nlmixr2.github.io/nonmem2rx/articles/simulate-new-dosing-with-covs.html). This example shows approaches to resample from the input dataset for covariate selection.
With nonmem2rx and babelmixr2, convert the imported rxode2 model to a nlmixr2 object, allowing:
- [Generation of Word and PowerPoint plots with
nlmixr2rpt](https://nlmixr2.github.io/nonmem2rx/articles/create-office.html)
- [Easy VPC
creation](https://nlmixr2.github.io/nonmem2rx/articles/create-vpc.html)
(with `vpcPlot()`)
- [Easy Individual plots with extra solved
points](https://nlmixr2.github.io/nonmem2rx/articles/create-augPred.html). This
will show the curvature of individual and population fits for
sparse data-sets (with `augPred()`)
You can even use this conversion to help debug your NONMEM model (or
even try it in nlmixr2 instead)
- [Understand how to simplify the NONMEM model to avoid rounding errors](https://nlmixr2.github.io/nonmem2rx/articles/read-rounding.html)
- [Run nlmixr2’s covariance step when NONMEMs covariance step has failed](https://nlmixr2.github.io/nonmem2rx/articles/read-rounding.html#step-5-get-the-covariance-of-the-model) (in the linked example, there was no covariance step because rounding errors)
## Simple example
Once `nonmem2rx` has been loaded, you simply type the location of the
nonmem control stream for the parser to start. For example:
```{r example}
library(nonmem2rx)
# First we need the location of the nonmem control stream Since we are
# running an example, we will use one of the built-in examples in
# `nonmem2rx`
ctlFile <- system.file("mods/cpt/runODE032.ctl", package="nonmem2rx")
# You can use a control stream or other file. With the development
# version of `babelmixr2`, you can simply point to the listing file
mod <- nonmem2rx(ctlFile, lst=".res", save=FALSE)
mod
```
You can see this automatically validates NONMEM and rxode2 outputs for
a couple of metrics.
# External projects that contributed to the tool's validation
The `nonmem2rx` tool was validated against:
- The `PsN` library test suite of NONMEM listings
(https://github.com/UUPharmacometrics/PsN/tree/master/test)
- The ddmore model scrapings
(https://github.com/dpastoor/ddmore_scraping).
- Models from NONMEM design tutorial Bauer 2021 https://doi.org/10.1002/psp4.12713
- Models from NONMEM tutorial 1 (Bauer 2019) https://doi.org/10.1002/psp4.12404
- Models from NONMEM tutorial 2 (Bauer 2019) https://doi.org/10.1002/psp4.12422
Due to the sheer size of the zipped models for these nonmem control
stream sources, these are excluded to keep the binary below 3 mgs
(CRAN requirement).
However, I would like to acknowledge all who helped in these projects.
With these projects the NONMEM conversion to rxode2 has been made much
more robust.
Still, while the tests are not/will not be in the CRAN binaries, you
can test them yourself by:
1. Downloading this repository
2. Running the tests `devtools::test()`