diff --git a/0.1/index.bs b/0.1/index.bs
index 8a64973c..b60204f1 100644
--- a/0.1/index.bs
+++ b/0.1/index.bs
@@ -13,6 +13,7 @@ Local Boilerplate: copyright yes
Boilerplate: style-darkmode off
Markup Shorthands: markdown yes
Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org
+Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org
Abstract: This document contains next-generation file format (NGFF)
Abstract: specifications for storing bioimaging data in the cloud.
Abstract: All specifications are submitted to the https://image.sc community for review.
@@ -98,6 +99,12 @@ is represented here as it would appear locally but could equally
be stored on a web server to be accessed via HTTP or in object storage
like S3 or GCS.
+Images {#image-layout}
+----------------------
+
+The following layout describes the expected Zarr hierarchy for images with
+multiple levels of resolutions and optionally associated labels.
+
```
. # Root folder, potentially in S3,
│ # with a flat list of images by image ID.
@@ -143,6 +150,50 @@ like S3 or GCS.
└── n
```
+High-content screening {#hcs-layout}
+------------------------------------
+
+The following specification defines the hierarchy for a high-content screening
+dataset. Three groups must be defined above the images:
+
+- the group above the images defines the well and MUST implement the
+ [well specification](#well-md). All images contained in a well are fields
+ of view of the same well
+- the group above the well defines a row of wells
+- the group above the well row defines an entire plate i.e. a two-dimensional
+ collection of wells organized in rows and columns. It MUST implement the
+ [plate specification](#plate-md)
+
+
+```
+. # Root folder, potentially in S3,
+│
+└── 5966.zarr # One plate (id=5966) converted to Zarr
+ ├── .zgroup
+ ├── .zattrs # Implements "plate" specification
+ ├── A # First row of the plate
+ │ ├── .zgroup
+ │ │
+ │ ├── 1 # First column of row A
+ │ │ ├── .zgroup
+ │ │ ├── .zattrs # Implements "well" specification
+ │ │ │
+ │ │ ├── 0 # First field of view of well A1
+ │ │ │ │
+ │ │ │ ├── .zgroup
+ │ │ │ ├── .zattrs # Implements "multiscales", "omero"
+ │ │ │ ├── 0
+ │ │ │ │ ... # Resolution levels
+ │ │ │ ├── n
+ │ │ │ └── labels # Labels (optional)
+ │ │ ├── ... # Fields of view
+ │ │ └── m
+ │ ├── ... # Columns
+ │ └── 12
+ ├── ... # Rows
+ └── H
+```
+
Metadata {#metadata}
====================
@@ -272,6 +323,193 @@ above).
]
```
+"plate" metadata {#plate-md}
+----------------------------
+
+For high-content screening datasets, the plate layout can be found under the
+custom attributes of the plate group under the `plate` key.
+
+
+ - acquisitions
+ - An optional list of JSON objects defining the acquisitions for a given
+ plate. Each acquisition object MUST contain an `id` key providing an
+ unique identifier within the context of the plate to which fields of
+ view can refer to. It SHOULD contain a `name` key identifying the name
+ of the acquisition. It SHOULD contain a `maximumfieldcount` key
+ indicating the maximum number of fields of view for the acquisition. It
+ MAY contain a `description` key providing a description for the
+ acquisition. It MAY contain a `startime` and/or `endtime` key specifying
+ the start and/or end timestamp of the acquisition using an epoch
+ string.
+ - columns
+ - A list of JSON objects defining the columns of the plate. Each column
+ object defines the properties of the column at the index of the object
+ in the list. If not empty, it MUST contain a `name` key specifying the
+ column name.
+ - field_count
+ - An integer defining the maximum number of fields per view across all
+ wells.
+ - name
+ - A string defining the name of the plate.
+ - rows
+ - A list of JSON objects defining the rows of the plate. Each row object
+ defines the properties of the row at the index of the object in the
+ list. If not empty, it MUST contain a `name` key specifying the row
+ name.
+ - version
+ - A string defining the version of the specification.
+ - wells
+ - A list of JSON objects defining the wells of the plate. Each well object
+ MUST contain a `path` key identifying the path to the well subgroup.
+
+
+For example the following JSON object defines a plate with two acquisition and
+6 wells (2 rows and 3 columns), containing up 2 fields of view per acquistion.
+
+```json
+"plate": {
+ "acquisitions": [
+ {
+ "id": 1,
+ "maximumfieldcount": 2,
+ "name": "Meas_01(2012-07-31_10-41-12)",
+ "starttime": 1343731272000
+ },
+ {
+ "id": 2,
+ "maximumfieldcount": 2,
+ "name": "Meas_02(201207-31_11-56-41)",
+ "starttime": 1343735801000
+ }
+ ],
+ "columns": [
+ {
+ "name": "1"
+ },
+ {
+ "name": "2"
+ },
+ {
+ "name": "3"
+ }
+ ],
+ "field_count": 4,
+ "name": "test",
+ "rows": [
+ {
+ "name": "A"
+ },
+ {
+ "name": "B"
+ }
+ ],
+ "version": "0.1",
+ "wells": [
+ {
+ "path": "2020-10-10/A/1"
+ },
+ {
+ "path": "2020-10-10/A/2"
+ },
+ {
+ "path": "2020-10-10/A/3"
+ },
+ {
+ "path": "2020-10-10/B/1"
+ },
+ {
+ "path": "2020-10-10/B/2"
+ },
+ {
+ "path": "2020-10-10/B/3"
+ }
+ ]
+ }
+```
+
+"well" metadata {#well-md}
+--------------------------
+
+For high-content screening datasets, the metadata about all fields of views
+under a given well can be found under the "well" key in the attributes of the
+well group.
+
+
+ - images
+ - A list of JSON objects defining the fields of views for a given well.
+ Each object MUST contain a `path` key identifying the path to the
+ field of view. If multiple acquisitions were performed in the plate, it
+ SHOULD contain an `acquisition` key identifying the id of the
+ acquisition which must match one of acquisition JSON objects defined in
+ the plate metadata.
+ - version
+ - A string defining the version of the specification.
+
+
+For example the following JSON object defines a well with four fields of
+views. The first two fields of view were part of the first acquisition while
+the last two fields of view were part of the second acquisition.
+
+```json
+"well": {
+ "images": [
+ {
+ "acquisition": 1,
+ "path": "0"
+ },
+ {
+ "acquisition": 1,
+ "path": "1"
+ },
+ {
+ "acquisition": 2,
+ "path": "2"
+ },
+ {
+ "acquisition": 2,
+ "path": "3"
+ }
+ ],
+ "version": "0.1"
+ }
+```
+
+Implementations {#implementations}
+==================================
+
+Projects which support reading and/or writing OME-NGFF data include:
+
+
+
+ - [omero-ms-zarr](https://github.com/ome/omero-ms-zarr)
+ - A microservice for OMERO.server that converts images stored in OMERO to OME Zarr files on the fly, served via a web API.
+
+ - [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools)
+ - A full workflow demonstrating the conversion of IDR images to OME Zarr images on S3.
+
+ - [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr)
+ - An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file.
+
+ - [ome-zarr-py](https://github.com/ome/ome-zarr-py)
+ - A napari plugin for reading ome-zarr files.
+
+ - [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw)
+ - A performant, Bio-Formats image file format converter.
+
+ - [vizarr](https://github.com/hms-dbmi/vizarr/)
+ - A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy.
+
+
+
+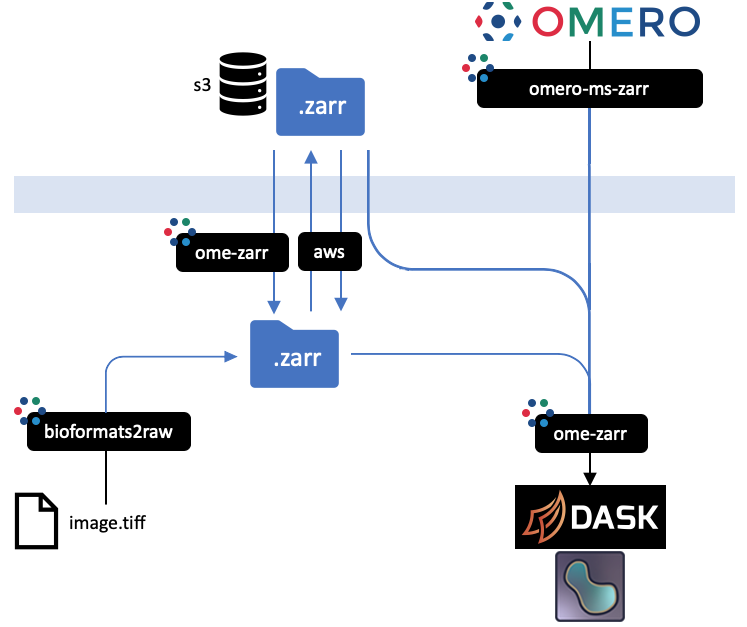 +
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
Citing {#citing}
================
@@ -293,24 +531,14 @@ Version History {#history}
+
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
Citing {#citing}
================
@@ -293,24 +531,14 @@ Version History {#history}
- | 0.1.3-dev4 |
- 2020-09-14 |
- Add the image-label object |
-
-
- | 0.1.3-dev3 |
- 2020-09-01 |
- Convert labels to multiscales |
+ 0.1.4 |
+ 2020-11-26 |
+ Add HCS specification |
- | 0.1.3-dev2 |
- 2020-08-18 |
- Rename masks as labels |
-
-
- | 0.1.3-dev1 |
- 2020-07-07 |
- Add mask metadata |
+ 0.1.3 |
+ 2020-09-14 |
+ Add labels specification |
| 0.1.2 |
diff --git a/latest/index.bs b/latest/index.bs
index 9dc7c4ff..31752fcb 100644
--- a/latest/index.bs
+++ b/latest/index.bs
@@ -14,6 +14,7 @@ Local Boilerplate: copyright yes
Boilerplate: style-darkmode off
Markup Shorthands: markdown yes
Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org
+Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org
Abstract: This document contains next-generation file format (NGFF)
Abstract: specifications for storing bioimaging data in the cloud.
Abstract: All specifications are submitted to the https://image.sc community for review.
@@ -100,6 +101,12 @@ is represented here as it would appear locally but could equally
be stored on a web server to be accessed via HTTP or in object storage
like S3 or GCS.
+Images {#image-layout}
+----------------------
+
+The following layout describes the expected Zarr hierarchy for images with
+multiple levels of resolutions and optionally associated labels.
+
```
. # Root folder, potentially in S3,
│ # with a flat list of images by image ID.
@@ -145,6 +152,50 @@ like S3 or GCS.
└── n
```
+High-content screening {#hcs-layout}
+------------------------------------
+
+The following specification defines the hierarchy for a high-content screening
+dataset. Three groups must be defined above the images:
+
+- the group above the images defines the well and MUST implement the
+ [well specification](#well-md). All images contained in a well are fields
+ of view of the same well
+- the group above the well defines a row of wells
+- the group above the well row defines an entire plate i.e. a two-dimensional
+ collection of wells organized in rows and columns. It MUST implement the
+ [plate specification](#plate-md)
+
+
+```
+. # Root folder, potentially in S3,
+│
+└── 5966.zarr # One plate (id=5966) converted to Zarr
+ ├── .zgroup
+ ├── .zattrs # Implements "plate" specification
+ ├── A # First row of the plate
+ │ ├── .zgroup
+ │ │
+ │ ├── 1 # First column of row A
+ │ │ ├── .zgroup
+ │ │ ├── .zattrs # Implements "well" specification
+ │ │ │
+ │ │ ├── 0 # First field of view of well A1
+ │ │ │ │
+ │ │ │ ├── .zgroup
+ │ │ │ ├── .zattrs # Implements "multiscales", "omero"
+ │ │ │ ├── 0
+ │ │ │ │ ... # Resolution levels
+ │ │ │ ├── n
+ │ │ │ └── labels # Labels (optional)
+ │ │ ├── ... # Fields of view
+ │ │ └── m
+ │ ├── ... # Columns
+ │ └── 12
+ ├── ... # Rows
+ └── H
+```
+
Metadata {#metadata}
====================
@@ -274,6 +325,157 @@ above).
]
```
+"plate" metadata {#plate-md}
+----------------------------
+
+For high-content screening datasets, the plate layout can be found under the
+custom attributes of the plate group under the `plate` key.
+
+
+ - acquisitions
+ - An optional list of JSON objects defining the acquisitions for a given
+ plate. Each acquisition object MUST contain an `id` key providing an
+ unique identifier within the context of the plate to which fields of
+ view can refer to. It SHOULD contain a `name` key identifying the name
+ of the acquisition. It SHOULD contain a `maximumfieldcount` key
+ indicating the maximum number of fields of view for the acquisition. It
+ MAY contain a `description` key providing a description for the
+ acquisition. It MAY contain a `startime` and/or `endtime` key specifying
+ the start and/or end timestamp of the acquisition using an epoch
+ string.
+ - columns
+ - A list of JSON objects defining the columns of the plate. Each column
+ object defines the properties of the column at the index of the object
+ in the list. If not empty, it MUST contain a `name` key specifying the
+ column name.
+ - field_count
+ - An integer defining the maximum number of fields per view across all
+ wells.
+ - name
+ - A string defining the name of the plate.
+ - rows
+ - A list of JSON objects defining the rows of the plate. Each row object
+ defines the properties of the row at the index of the object in the
+ list. If not empty, it MUST contain a `name` key specifying the row
+ name.
+ - version
+ - A string defining the version of the specification.
+ - wells
+ - A list of JSON objects defining the wells of the plate. Each well object
+ MUST contain a `path` key identifying the path to the well subgroup.
+
+
+For example the following JSON object defines a plate with two acquisition and
+6 wells (2 rows and 3 columns), containing up 2 fields of view per acquistion.
+
+```json
+"plate": {
+ "acquisitions": [
+ {
+ "id": 1,
+ "maximumfieldcount": 2,
+ "name": "Meas_01(2012-07-31_10-41-12)",
+ "starttime": 1343731272000
+ },
+ {
+ "id": 2,
+ "maximumfieldcount": 2,
+ "name": "Meas_02(201207-31_11-56-41)",
+ "starttime": 1343735801000
+ }
+ ],
+ "columns": [
+ {
+ "name": "1"
+ },
+ {
+ "name": "2"
+ },
+ {
+ "name": "3"
+ }
+ ],
+ "field_count": 4,
+ "name": "test",
+ "rows": [
+ {
+ "name": "A"
+ },
+ {
+ "name": "B"
+ }
+ ],
+ "version": "0.1",
+ "wells": [
+ {
+ "path": "2020-10-10/A/1"
+ },
+ {
+ "path": "2020-10-10/A/2"
+ },
+ {
+ "path": "2020-10-10/A/3"
+ },
+ {
+ "path": "2020-10-10/B/1"
+ },
+ {
+ "path": "2020-10-10/B/2"
+ },
+ {
+ "path": "2020-10-10/B/3"
+ }
+ ]
+ }
+```
+
+"well" metadata {#well-md}
+--------------------------
+
+For high-content screening datasets, the metadata about all fields of views
+under a given well can be found under the "well" key in the attributes of the
+well group.
+
+
+ - images
+ - A list of JSON objects defining the fields of views for a given well.
+ Each object MUST contain a `path` key identifying the path to the
+ field of view. If multiple acquisitions were performed in the plate, it
+ SHOULD contain an `acquisition` key identifying the id of the
+ acquisition which must match one of acquisition JSON objects defined in
+ the plate metadata.
+ - version
+ - A string defining the version of the specification.
+
+
+For example the following JSON object defines a well with four fields of
+views. The first two fields of view were part of the first acquisition while
+the last two fields of view were part of the second acquisition.
+
+```json
+"well": {
+ "images": [
+ {
+ "acquisition": 1,
+ "path": "0"
+ },
+ {
+ "acquisition": 1,
+ "path": "1"
+ },
+ {
+ "acquisition": 2,
+ "path": "2"
+ },
+ {
+ "acquisition": 2,
+ "path": "3"
+ }
+ ],
+ "version": "0.1"
+ }
+```
+
Implementations {#implementations}
==================================
@@ -331,24 +533,14 @@ Version History {#history}
- | 0.1.3-dev4 |
- 2020-09-14 |
- Add the image-label object |
-
-
- | 0.1.3-dev3 |
- 2020-09-01 |
- Convert labels to multiscales |
+ 0.1.4 |
+ 2020-11-26 |
+ Add HCS specification |
- | 0.1.3-dev2 |
- 2020-08-18 |
- Rename masks as labels |
-
-
- | 0.1.3-dev1 |
- 2020-07-07 |
- Add mask metadata |
+ 0.1.3 |
+ 2020-09-14 |
+ Add labels specification |
| 0.1.2 |
 +
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
Citing {#citing}
================
@@ -293,24 +531,14 @@ Version History {#history}
+
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
Citing {#citing}
================
@@ -293,24 +531,14 @@ Version History {#history}