NeuronUnit: A SciUnit repository for neuroscience-related tests, capabilities and so on.
https://github.com/rgerkin/papers/blob/master/neuronunit_frontiers/Paper.pdf
INCF Meeting (August, 2014) (Less code) https://github.com/scidash/assets/blob/master/presentations/SciUnit%20INCF%20Talk.pdf?raw=true
OpenWorm Journal Club (August, 2014) (More code) https://github.com/scidash/assets/blob/master/presentations/SciUnit%20OpenWorm%20Journal%20Club.pdf?raw=true
from channel_worm.ion_channel.models import GraphData
from neuronunit.tests.channel import IVCurvePeakTest
from neuronunit.models.channel import ChannelModel
# Instantiate the model
channel_model_name = 'EGL-19.channel' # Name of a NeuroML channel model
channel_id = 'ca_boyle'
channel_file_path = os.path.join('path', 'to', 'models', '%s.nml' % channel_model_name)
model = ChannelModel(channel_file_path, channel_index=0, name=channel_model_name)
# Get the experiment data from ChannelWorm and instantiate the test
doi = '10.1083/jcb.200203055'
fig = '2B'
sample_data = GraphData.objects.get(graph__experiment__reference__doi=doi,
graph__figure_ref_address=fig)
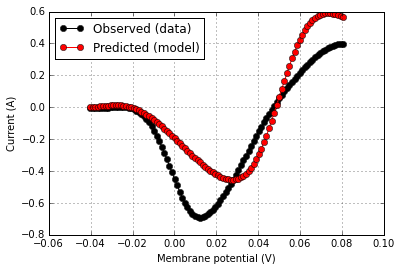
voltage, current_per_farad = sample_data.asunitedarray()
patch_capacitance = pq.Quantity(1e-13,'F') # Assume recorded patch had this capacitance;
# an arbitrary scaling factor.
current = current_per_farad * patch_capacitance
observation = {'v':voltage,
'i':current}
test = IVCurvePeakTest(observation)
# Judge the model output against the experimental data
score = test.judge(model)
rd = score.related_data
score.plot(rd['v'],rd['i_obs'],color='k',label='Observed (data)')
score.plot(rd['v'],rd['i_pred'],same_fig=True,color='r',label='Predicted (model)')score.summarize()
""" OUTPUT:
Model EGL-19.channel (ChannelModel) achieved score Fail on test 'IV Curve Test (IVCurvePeakTest)'. ===
"""
score.describe()
""" OUTPUT:
The score was computed according to 'The sum-squared difference in the observed and predicted current values over the range of the tested holding potentials.' with raw value 3.151 pA^2
"""
(Example 2) Testing the membrane potential and action potential widths of several spiking neuron models against experimental data found at http://neuroelectro.org.
import sciunit
from neuronunit import neuroelectro,tests,capabilities
import neuronunit.neuroconstruct.models as nc_models
from pythonnC.utils.putils import OSB_MODELS
# We will test cerebellar granule cell models.
brain_area = 'cerebellum'
neuron_type = 'cerebellar_granule_cell'
path = os.path.join(OSB_MODELS,brain_area,neuron_type)
neurolex_id = 'nifext_128' # Cerebellar Granule Cell
# Specify reference data for a test of resting potential for a granule cell.
reference_data = neuroelectro.NeuroElectroSummary(
neuron = {'nlex_id':neurolex_id}, # Neuron type.
ephysprop = {'name':'Resting Membrane Potential'}) # Electrophysiological property name.
# Get and verify summary data for the combination above from neuroelectro.org.
reference_data.get_values()
vm_test = tests.RestingPotentialTest(
observation = {'mean':reference_data.mean,
'std':reference_data.std},
name = 'Resting Potential')
# Specify reference data for a test of action potential width.
reference_data = neuroelectro.NeuroElectroSummary(
neuron = {'nlex_id':neurolex_id}, # Neuron type.
ephysprop = {'name':'Spike Half-Width'}) # Electrophysiological property name.
# Get and verify summary data for the combination above from neuroelectro.org.
reference_data.get_values()
spikewidth_test = tests.InjectedCurrentSpikeWidthTest(
observation = {'mean':reference_data.mean,
'std':reference_data.std},
name = 'Spike Width',
params={'injected_current':{'ampl':0.0053}}) # 0.0053 nA (5.3 pA) of injected current.
# Create a test suite from these two tests.
suite = sciunit.TestSuite('Tests',(spikewidth_test,vm_test))
models = []
for model_name in model_names # Iterate through a list of models downloaded from http://opensourcebrain.org
model_info = (brain_area,neuron_type,model_name)
model = nc_models.OSBModel(*model_info)
models.append(model) # Add to the list of models to be tested.
score_matrix = suite.judge(models,stop_on_error=True)
score_matrix.view()
| Model | Spike Width | Resting Potential |
|---|---|---|
| (InjectedCurrentSpikeWidthTest) | (RestingPotentialTest) | |
| cereb_grc_mc (OSBModel) | Z = nan | Z = 4.92 |
| GranuleCell (OSBModel) | Z = -3.56 | Z = 0.88 |
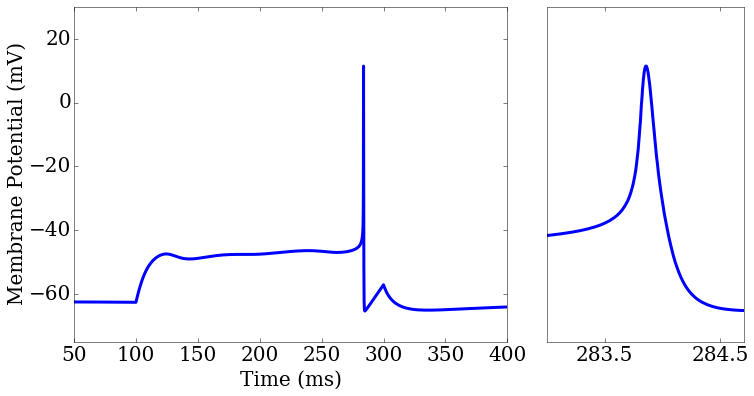
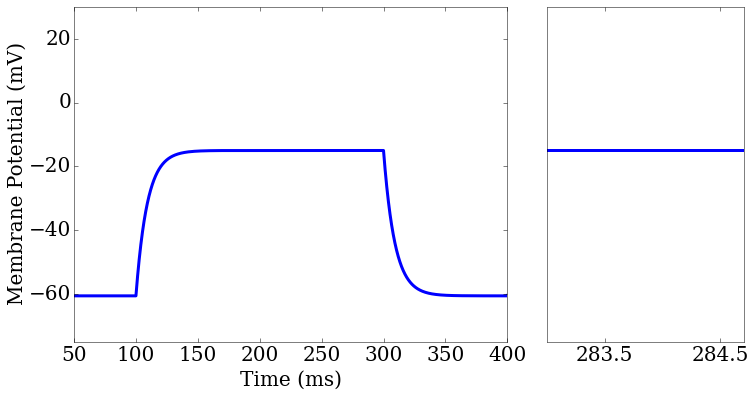
import matplotlib.pyplot as plt
ax1 = plt.subplot2grid((1,3), (0,0), colspan=2)
ax2 = plt.subplot2grid((1,3), (0,2), colspan=1)
rd = score.related_data
score_matrix[0,0].plot(rd['t'],rd['v_pred'],ax=ax1[0])
score_matrix[0,1].plot(rd['t'],rd['v_pred'],ax=ax2[0])
ax2.set_xlim(283,284.7)