-
Notifications
You must be signed in to change notification settings - Fork 0
Home
MGSM is a BEAST 2 package, so you need to get BEAST 2 first.
To run an multi-gamma site model analysis, you need the MGSM package. The easiest way to do this is by starting BEAUti, and open the package manager using the File/Mange packages menu. A dialog pops up that looks like this:
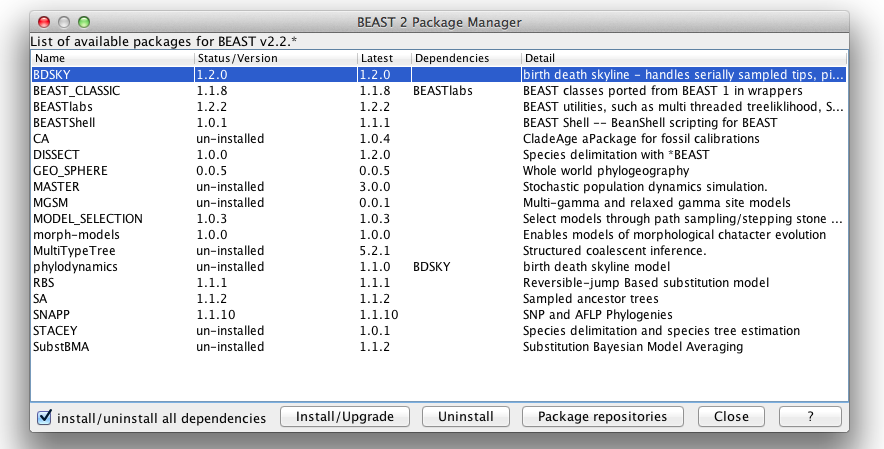
Select the MGSM package and click the Install/Upgrade button. Restart BEAUti in order for the multi-gamma site model to be available.
Start BEAUti, load an alignment as per usual. Now, when you go to the Site model panel, there is a choice of site models shown in the combobox at the top of the panel:
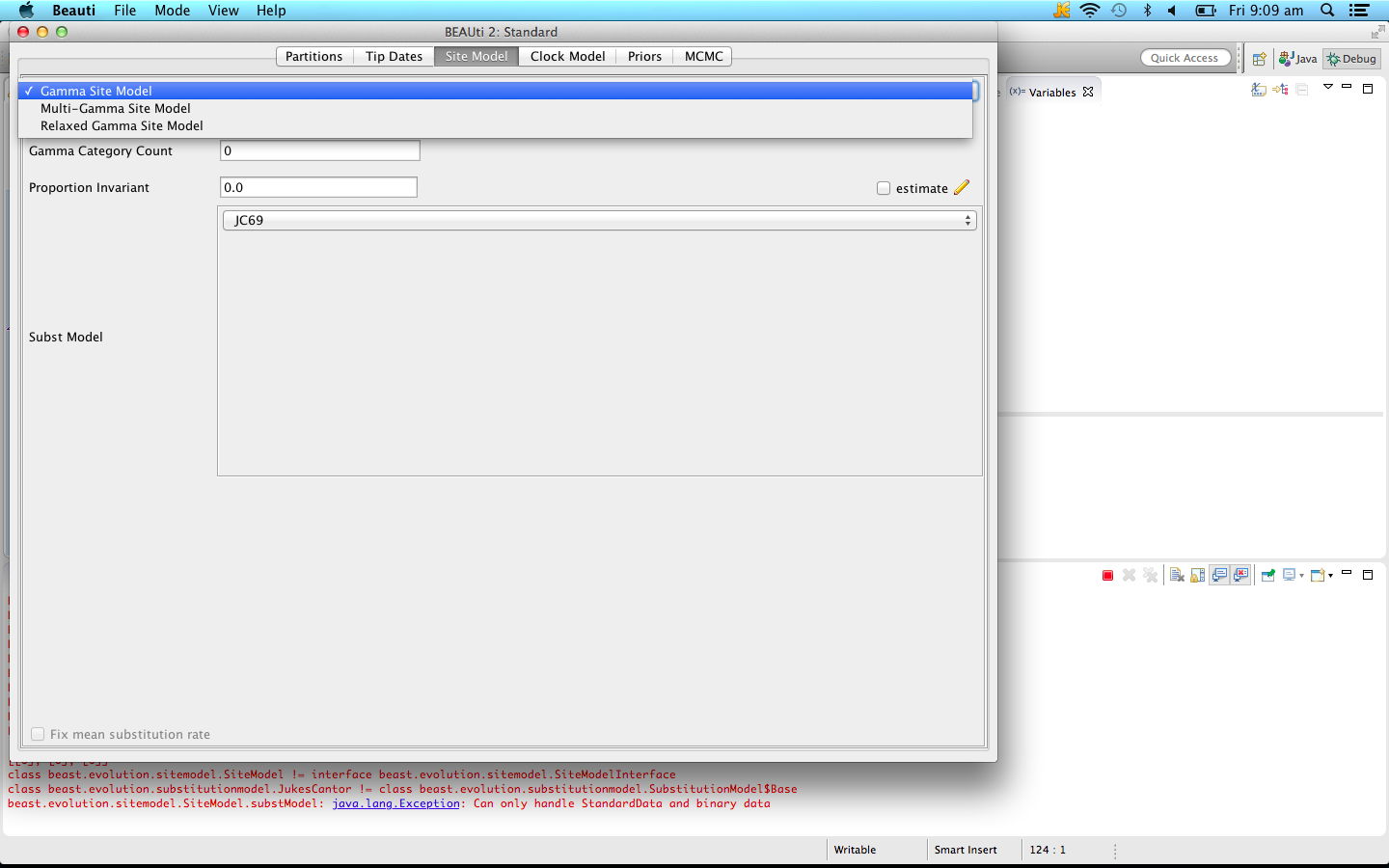
Select the Multi-Gamma Site Model for the model where each branch has independently selected gamma shape parameters. Select the Relaxed Gamma Site Model for models where the shape parameter is drawn from a log-normal distribution (recommended).
You can now select whether you want to estimate the proportion invariant, the relative substitution rate (if you have multiple partitions on the same tree) and the number of gamma categories. If you have a significant number of constant sites, it makes sense to estimate the proportion invariant. Failing to do so causes the slowest gamma category to cover the invariant sites, and results in a highly distorted gamma shape to accommodate these constant sites.
First, you want to run your analysis with the gamma site model. Higher shape parameters estimates mean lower rate variation. Shape parameters over 16 indicate there is virtually no rate variation at all. If the estimate of the shape parameter is high (> 5 or so) it probably does not make sense to investigate more sophisticated site models. If the shape parameter is very low (<0.1) either you need to use invariant sites, or increase the number of rate categories.
Once you have run a single gamma site model analysis, you can run a relaxed gamma site model analysis. The first thing to compare after convergence is the marginal likelihoods of the relaxed and single gamma site models. If the mean is higher for the relaxed site model and the 95% HPDs of the marginal likelihoods do not overlap in Tracer, the relaxed gamma site model fits better. If the distributions overlaps, you need a stricter test. Using the AICM for model comparison (menu Analysis/Model Comparison in Tracer) is desirable over the harmonic mean (HME). Path sampling (see model-selection package in BEAST) is even better.
The MGSM model reports gamma shapes for each branch in the tree, but these branches can move around during the MCMC: only the branches for the leaves can be guaranteed to be linked to the same gamma shape and are numbered 0 to n-1 for a tree with n taxa (the root is also fixed at 2n-2, but that shape is not used in the inference).
So, what you should see in a tracelog that is properly mixed is good ESSs for (most of) the first n shape parameters (for a partition). The ESSs for the following n-2 shape parameters are meaningless because they cannot be guaranteed to be associated with the same clade, and are therefore a mixture of distributions. Low ESSs in these shape parameters is no reason for concern.
If you have questions, please use the BEAST users mailing list (in google groups or mark mail).
A more detailed description of the method can be found in Remco Bouckaert and Peter Lockhart. "Heterotachy and multi gamma site models", BioArxiv, 2015.