-
Notifications
You must be signed in to change notification settings - Fork 40
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Strip outputs from .ipynb files [skip ci]
- Loading branch information
1 parent
248c308
commit 6e299b8
Showing
30 changed files
with
11,207 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,200 @@ | ||
| --- | ||
| title: An R Markdown document converted from "AC1/r-proxy-controls.irnb" | ||
| output: html_document | ||
| --- | ||
|
|
||
| # Negative (Proxy) Controls for Unobserved Confounding | ||
|
|
||
| Consider the following SEM, where $Y$ is the outcome, $D$ is the treatment, $A$ is some unobserved confounding, and $Q$, $X$, $S$ are the observed covariates. In particular, $Q$ is considered to be the proxy control treatment as it a priori has no effect on the actual outcome $Y$, and $S$ is considered to be the proxy control outcome as it a priori is not affected by the actual treatment $D$. See also [An Introduction to Proximal Causal Learning](https://arxiv.org/pdf/2009.10982.pdf), for more information on this setting. | ||
|
|
||
| 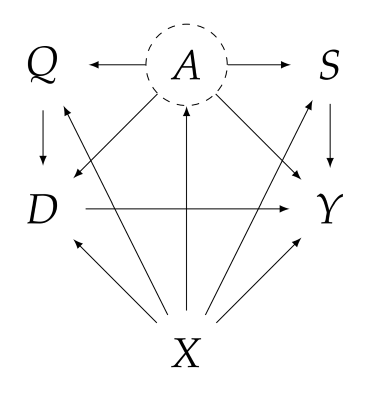 | ||
|
|
||
| Under linearity assumptions, the average treatment effect can be estimated by solving the vector of moment equations: | ||
| \begin{align} | ||
| E\left[(\tilde{Y} - \alpha \tilde{D} - \delta \tilde{S}) \left(\begin{aligned}\tilde{D}\\ \tilde{Q}\end{aligned}\right) \right] = 0 | ||
| \end{align} | ||
| where for every variable $V$ we denote with $\tilde{V} = V - E[V|X]$. | ||
|
|
||
| When the dimension of the proxy treatment variables $Q$ is larger than the dimension of proxy outcome variables $S$, then the above system of equations is over-identified. In these settings, we first project the "technical instrument" variables $\tilde{V}=(\tilde{D}, \tilde{Q})$ onto the space of "technical treatment" variables $\tilde{W}=(\tilde{D}, \tilde{S})$ and use the projected $\tilde{V}$ as a new "technical instrument". In particular, we run an OLS regression of $\tilde{W}$ on $\tilde{V},$ and define $\tilde{Z} = E[\tilde{W}\mid \tilde{V}] = B \tilde{V}$, where the $t$-th row $\beta_t$ of the matrix $B$ is the OLS coefficient in the regression of $\tilde{W}_t$ on $\tilde{V}$. These new variables $\tilde{Z}$, can also be viewed as engineered technical instrumental variables. Then we have the exactly identified system of equations: | ||
| \begin{align} | ||
| E\left[(\tilde{Y} - \alpha \tilde{D} - \delta \tilde{S}) \tilde{Z} \right] := E\left[(\tilde{Y} - \alpha \tilde{D} - \delta \tilde{S}) B \left(\begin{aligned}\tilde{D}\\ \tilde{Q}\end{aligned}\right) \right] = 0 | ||
| \end{align} | ||
|
|
||
| The solution to this system of equations is numerically equivalent to the following two stage algorithm: | ||
| - Run OLS of $\tilde{W}=(\tilde{D}, \tilde{S})$ on $\tilde{V}=(\tilde{D}, \tilde{Q})$ | ||
| - Define $\tilde{Z}$ as the predictions of the OLS model | ||
| - Run OLS of $\tilde{Y}$ on $\tilde{Z}$. | ||
| This is the well-known Two-Stage-Least-Squares (2SLS) algorithm for instrumental variable regression. | ||
|
|
||
| Since we're considering only linear models and in a low-dimensional setting, we'll focus on just using linear IV methods. | ||
|
|
||
| ```{r} | ||
| install.packages("hdm") | ||
| ``` | ||
|
|
||
| ```{r} | ||
| library(hdm) | ||
| set.seed(1) | ||
| ``` | ||
|
|
||
| # Analyzing Simulated Data | ||
|
|
||
| First, let's evaluate the methods on simulated data generated from a linear SEM characterized by the above DAG. For this simulation, we'll set the ATE to 2. | ||
|
|
||
| ```{r} | ||
| gen_data <- function(n, ate) { | ||
| X <- matrix(rnorm(n * 10), ncol = 10) | ||
| A <- 2 * X[, 1] + rnorm(n) | ||
| Q <- 10 * A + 2 * X[, 1] + rnorm(n) | ||
| S <- 5 * A + X[, 1] + rnorm(n) | ||
| D <- Q - A + 2 * X[, 1] + rnorm(n) | ||
| Y <- ate * D + 5 * A + 2 * S + 0.5 * X[, 1] + rnorm(n) | ||
| return(list(X, A, Q, S, D, Y)) | ||
| } | ||
| ``` | ||
|
|
||
| ```{r} | ||
| data_list <- gen_data(5000, 2) | ||
| X <- data_list[[1]] | ||
| A <- data_list[[2]] | ||
| Q <- data_list[[3]] | ||
| S <- data_list[[4]] | ||
| D <- data_list[[5]] | ||
| Y <- data_list[[6]] | ||
| ``` | ||
|
|
||
| We define the technical instrument $V=(D, Q)$ and technical treatment $W=(D, S)$. Estimating the treatement effect is then just a matter of solving an instrument variable regression problem with instruments $V$ and treatments $W$ and looking at the first coefficient associated with $D$. | ||
|
|
||
| ```{r} | ||
| W <- cbind(D, S) | ||
| V <- cbind(D, Q) | ||
| ``` | ||
|
|
||
| ```{r} | ||
| piv <- tsls(X, W, Y, V, homoscedastic = FALSE) | ||
| cat("Estimated coefficient:", piv$coefficients["D", 1], "\n") | ||
| cat("Standard error:", piv$se["D"], "\n") | ||
| ``` | ||
|
|
||
| # With Cross-Fitting | ||
|
|
||
| We can also consider partialling out the controls using DML with cross-fitting | ||
|
|
||
| ```{r} | ||
| lm_dml_for_proxyiv <- function(x, d, q, s, y, dreg, qreg, yreg, sreg, nfold = 5) { | ||
| # this implements DML for a partially linear IV model | ||
| nobs <- nrow(x) | ||
| foldid <- rep.int(1:nfold, times = ceiling(nobs / nfold))[sample.int(nobs)] | ||
| I <- split(1:nobs, foldid) | ||
| # create residualized objects to fill | ||
| ytil <- dtil <- qtil <- stil <- rep(NA, nobs) | ||
| # obtain cross-fitted residuals | ||
| cat("fold: ") | ||
| for (b in seq_along(I)) { | ||
| dfit <- dreg(x[-I[[b]], ], d[-I[[b]]]) # take a fold out | ||
| qfit <- qreg(x[-I[[b]], ], q[-I[[b]]]) # take a fold out | ||
| yfit <- yreg(x[-I[[b]], ], y[-I[[b]]]) # take a fold out | ||
| sfit <- sreg(x[-I[[b]], ], s[-I[[b]]]) # take a fold out | ||
| dtil[I[[b]]] <- (d[I[[b]]] - x[I[[b]], ] %*% as.matrix(dfit$coefficients)) # record residual | ||
| qtil[I[[b]]] <- (q[I[[b]]] - x[I[[b]], ] %*% as.matrix(qfit$coefficients)) # record residual | ||
| ytil[I[[b]]] <- (y[I[[b]]] - x[I[[b]], ] %*% as.matrix(yfit$coefficients)) # record residial | ||
| stil[I[[b]]] <- (s[I[[b]]] - x[I[[b]], ] %*% as.matrix(sfit$coefficients)) # record residual | ||
| cat(b, " ") | ||
| } | ||
| ivfit <- tsls(y = ytil, d = cbind(dtil, stil), x = NULL, z = cbind(dtil, qtil), | ||
| intercept = FALSE, homoscedastic = FALSE) | ||
| coef_est <- ivfit$coef[1] # extract coefficient | ||
| se <- ivfit$se[1] # record standard error | ||
| cat(sprintf("\ncoef (se) = %g (%g)\n", coef_est, se)) | ||
| return(list(coef_est = coef_est, se = se, dtil = dtil, qtil = qtil, | ||
| ytil = ytil, stil = stil, foldid = foldid, spI = I)) | ||
| } | ||
| ``` | ||
|
|
||
| We'll just use OLS for partialling out again. We could of course try something more elaborate if we wanted. | ||
|
|
||
| ```{r} | ||
| dreg <- function(x, d) { | ||
| lm.fit(x, d) | ||
| } # ML method=ols | ||
| qreg <- function(x, q) { | ||
| lm.fit(x, q) | ||
| } # ML method=ols | ||
| yreg <- function(x, y) { | ||
| lm.fit(x, y) | ||
| } # ML method=ols | ||
| sreg <- function(x, s) { | ||
| lm.fit(x, s) | ||
| } # ML method=ols | ||
| dml_piv <- lm_dml_for_proxyiv(X, D, Q, S, Y, dreg, qreg, yreg, sreg, nfold = 5) | ||
| dml_piv | ||
| ``` | ||
|
|
||
| ## Real Data - Effects of Smoking on Birth Weight | ||
|
|
||
| In this study, we will be studying the effects of smoking on baby weight. We will consider the following stylized setup: | ||
|
|
||
| Outcome ($Y$): baby weight | ||
|
|
||
| Treatment ($D$): smoking | ||
|
|
||
| Unobserved confounding ($A$): family income | ||
|
|
||
| The observed covariates are put in to 3 groups: | ||
|
|
||
|
|
||
| * Proxy treatment control ($Q$): mother's education | ||
| * Proxy outcome control ($S$): parity (total number of previous pregnancies) | ||
| * Other observed covariates ($X$): mother's race and age | ||
|
|
||
|
|
||
| Education serves as a proxy treatment control $Q$ because it reflects unobserved confounding due to household income $A$ but has no direct medical effect on birth weight $Y$. Parity serves as a proxy outcome control $S$ because family size reflects household income $A$ but is not directly caused by smoking $D$ or education $Q$. | ||
|
|
||
| A description of the data used can be found [here](https://www.stat.berkeley.edu/users/statlabs/data/babies.readme). | ||
|
|
||
| ```{r} | ||
| data <- read.table("https://www.stat.berkeley.edu/users/statlabs/data/babies23.data", header = TRUE) | ||
| summary(data) | ||
| ``` | ||
|
|
||
| ```{r} | ||
| # Filter data to exclude entries where income, number of cigarettes smoked, | ||
| # race, age are not asked or not known | ||
| data <- data[data$race != 99, ] | ||
| data <- data[!(data$number %in% c(98, 99)), ] | ||
| data <- data[!(data$inc %in% c(98, 99)), ] | ||
| data <- data[data$age != 99, ] | ||
| dim(data) | ||
| ``` | ||
|
|
||
| ```{r} | ||
| # Create matrices for X, D, Q, S, A, Y | ||
| X <- model.matrix(~ 0 + C(race) + age, data) | ||
| D <- model.matrix(~ 0 + number, data) | ||
| Q <- model.matrix(~ 0 + ed, data) | ||
| S <- model.matrix(~ 0 + parity, data) | ||
| A <- model.matrix(~ 0 + inc, data) | ||
| Y <- model.matrix(~ 0 + wt, data) | ||
| ``` | ||
|
|
||
| ```{r} | ||
| # Use cross-fitting with OLS to estimate treatment effect within linear model context | ||
| dreg <- function(x, d) { | ||
| lm.fit(x, d) | ||
| } # ML method=ols | ||
| qreg <- function(x, q) { | ||
| lm.fit(x, q) | ||
| } # ML method=ols | ||
| yreg <- function(x, y) { | ||
| lm.fit(x, y) | ||
| } # ML method=ols | ||
| sreg <- function(x, s) { | ||
| lm.fit(x, s) | ||
| } # ML method=ols | ||
| dml_bw_piv <- lm_dml_for_proxyiv(X, D, Q, S, Y, dreg, qreg, yreg, sreg, nfold = 5) | ||
| dml_bw_piv | ||
| ``` | ||
|
|
Oops, something went wrong.