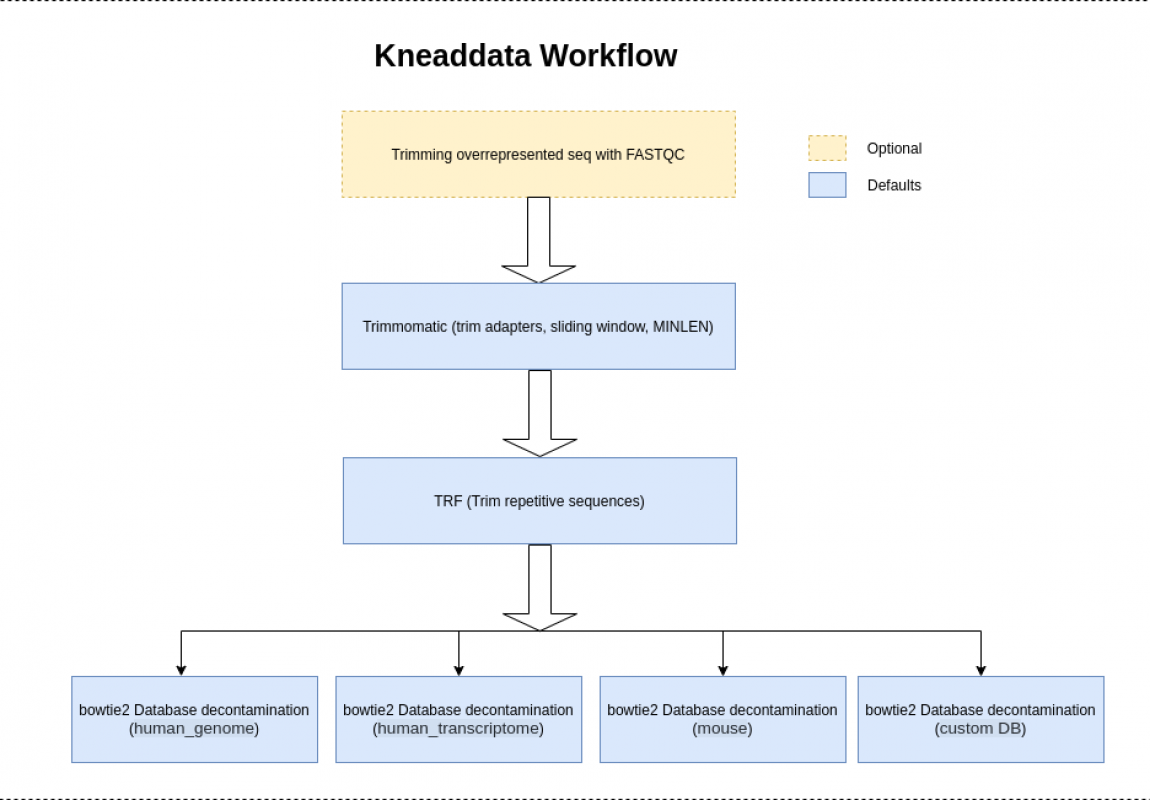
Nextflow workflow used to run kneaddata
The KneadData utility is used to preprocess metagenomic data for microbiome analysis by removing contaminating host sequences and running quality trimming.
A selection of reference database is provided so that the appropriate host genome can be used for decontamination.
Four types of output files will be created (where $INPUTNAME is the basename of $INPUT):
- The final file of filtered sequences after trimming
- $OUTPUT_DIR/$INPUTNAME_kneaddata.fastq
- The contaminant sequences from testing against a database
- $OUTPUT_DIR/$INPUTNAME_kneaddata_$DATABASE_$SOFTWARE_contam.fastq
- The log file from the run
- $OUTPUT_DIR/$INPUTNAME_kneaddata.log
- The FASTQ file of trimmed sequences
-
$OUTPUT_DIR/$INPUTNAME_kneaddata.trimmed.fastq
-
Trimmomatic is run with the following arguments by default “SLIDINGWINDOW:4:20 MINLEN:70”. The minimum length is computed as 70 percent of the length of the input reads.