-
Notifications
You must be signed in to change notification settings - Fork 13
FAQs
I'm getting warnings about missing strain mappings / I don't understand how strain_mappings.csv is supposed to work
NPLinker can't automatically determine equivalence between strain labels from metabolomics data and strain labels from genomics data because they typically vary widely in form between and across datasets. To solve this problem, NPLinker expects to find a file called strain_mappings.csv supplied as part of any new dataset. As the name suggests this is a simple CSV format file which should have one line for each strain in the dataset. The first column should contain the name of the strain, followed by any other names used for the same strain. Internally, NPLinker will treat any occurrence of any of the names for the strain as equivalent.
For example, a very short strain_mappings.csv might contain these 3 lines:
strain1,strain1A,strain1.B,strain1_C,strainONE
strain2,strainTWO,strainTWO_
strain3
Taking this line by line, it is saying:
- strain1 is also known as strain1A, strain1.B, strain1_C, and strainONE. NPLinker will therefore treat any instances of the latter 4 labels as equivalent to strain1
- strain2 is also known as strainTWO and strainTWO_, so again NPLinker will treat these as equivalent
- strain3 is only known as strain3 in the dataset, it has no other labels
Each line of the file may have any number of columns, and as shown in the example above this number does NOT need to be consistent.
Since it may be difficult to determine if you have added all the necessary mappings, NPLinker will generate a pair of files called unknown_strains_met.csv and unknown_strains_gen.csv in the dataset folder each time it is run. These files contain any unknown strain labels encountered while parsing the metabolomics and genomics data respectively.
NPLinker is still showing lots of warnings about unknown strains even after adding strain_mappings.csv
This probably indicates NPLinker is unable to parse the strain labels from the BiG-SCAPE output files correctly (these labels are usually linked to the filenames of the antiSMASH .gbk files used as input to BiG-SCAPE).
By default, NPLinker attempts to parse the labels by extracting the text up to the first "." character. If that doesn't produce a recognised strain name it tries again, extracting the text up to the first "_". If that fails, it tries a final time by extracting the text up to the first "-". If none of these attempts produce a recognised strain name, it displays a warning message. If this is happening, you will likely find a long list of the affected names in unknown_strains_gen.csv in your dataset folder.
If your strain labels are not being parsed correctly by any of these 3 rules, you can modify or replace the rules through the nplinker.toml configuration file. Start by copying/pasting in the following content, which reflects the default behaviour:
[antismash]
antismash_delimiters = [".", "_", "-"]
As an example where the defaults won't work, take the label "S.collinus365.region003". This should become "S.collinus365" after parsing, but NPLinker will first parse it as simply "S" (up to the first ".") and then "S.collinus365.region003" because it doesn't contain a "_" or a "-". In a situation like this, you can modify the antismash_delimiters option to search for all text up to ".region" instead:
[antismash]
antismash_delimiters = [".region"]
Now when you run NPLinker, the application will search for ".region" in "S.collinus365.region003", find a match, remove all text up to that location, and give the result "S.collinus365" as desired.
When displaying scoring results, NPLinker will highlight strains that are shared between linked objects in green. Strains that are not shared will have a grey background. In the image below, CNS237 is shared between the GCF and the Spectrum it has been linked with, while CNT003 is only present in the GCF.

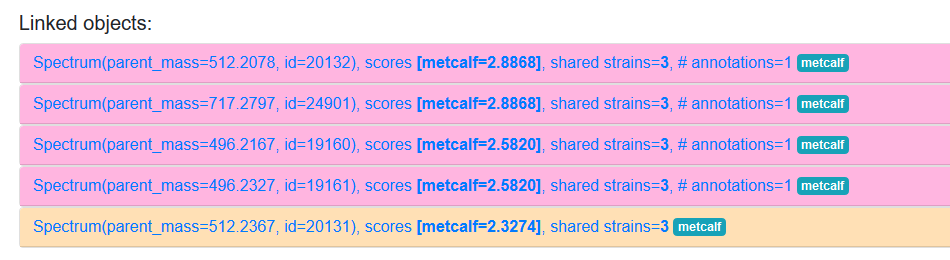
Normally lists of objects in the results will have a light orange background colour, like the final entry in the image below. However in some cases you may see a pinkish colour instead, as with the other resutls in the image. This simply indicates that the object in question has one or more annotations associated with it (e.g. from GNPS), which can be viewed by expanding the entry for the object.
A: Yes, simply click the CSV button below the table you want to export and your browser should prompt you to download a CSV file. This will be formatted to match the current state of the table, containing only the visible rows and having the same columns as are shown in the application.
Yes - this might be useful if you have a fast PC and a relatively slow laptop, and/or you have datasets stored on a server that would be difficult to copy to your own machine. To do this, you need to provide an extra parameter when running the Docker image that allows network connections from sources other than the local machine. The modified Docker command is:
docker run --name webapp -p 5006:5006 -v <shared folder>:/data:rw -e BOKEH_ALLOW_WS_ORIGIN=<IP address>:5006 andrewramsay/nplinker
where <IP address> should be replaced with the current IP address of the machine running Docker.