FIELDimageR-QGIS: A Tool to Analyze Images From Agricultural Field Trials in QGIS.
This tutorial is a compilation of functions made on R by Popat Pawar and Filipe Matias to analyze orthomosaic images from research fields using QGIS. The tutorial teaches how to build a plot shapefile to extract information for each plot, enabling the evaluation of different reflectance values at various wavelengths, vegetation indices, canopy percentage, counting plants (stand count), and plant height. It also guides in removing soil and segmenting orthomosaics to create masks.
Step 2. Loading mosaics and visualizing
Step 3. Building the plot shapefile
Step 4. Editing the plot shapefile and raster (e.g. move, rotate, scale, etc.)
Step 5. Building vegetation indices
Step 6. Removing the soil effect based on image segmentation
Step 7. Extracting data from field images and visualization
Step 9. Estimating object area percentage (e.g. canopy and LAI)
Step 10. Counting the number of objects and taking measurements (e.g. stand count, plants, etc.)
Step 12. Cropping individual plots and saving
Step 13. Image segmentation based on Kmeans (Unsupervised)
Step 14. Image segmentation based on machine learning models as Random Forest (Supervised)
Step 15. Area Under the Curve (AUC)
Step 16. Multispectral (merge multiple raster into a new raster)
Start the pipeline by installing the software:
- Attention: The first time running FIELDimageR-QGIS may takes time for installing all R packages!
Make sure to open the Processing Toolbox visualization in QGIS:
- (1) View
- (2) Panels
- (3) Processing Toolbox
- (4) Check if the rocessing Toolbox is showing in the right
The nest step is installing Processing R Provider plugin in QGIS following the steps:
- (1) Plugins
- (2) Manage and Install Plugins
- (3) All
- (4) Search for "Processing R Provider"
- (5) Install Plugin
- (6) Check for R at the Processing Toolbox
Downloading FIELDimageR-QGIS functions and saving on your rscripts QGIS folder:
Install - Option 01:
- (1) Link: https://github.com/filipematias23/FIELDimageR-QGIS
- (2) Press Code
- (3) Download ZIP
- (4) Unzip and save the functions from rscripts to your QGIS rscrits
- (5) If you don´t know how to open QGIS rscrits just go to QGIS > Processing Toolbox > Options (wrench icon)
- (6) On Providers Click on R
- (7) Copy the path at R scripts folder and open it on your explorer
- (8) Now past the copied FIELDimageR functions downloaded on rscripts folder from the GitHub Tutorial.
Install - Option 02:
Another easier option is adding the downloaded FIELDimageR-QGIS > rscripts folder as an additional rscripts at QGIS by following the steps below:
There are many ways to create or start a New Project in QGIS. The image below highlights some examples:
Uploading files (e.g., raster, table, shapefiles, etc.) to QGIS is very simple:
FIELDimageR-QGIS allows drawing the plot shape file using the function fieldShape at the Processing Toolbox > R > FIELDimageR > fieldShape to creat a basic grid_shapefile.
Installing QuickMapServices plugin in QGIS following the steps:
Plugins menu toolbar > Manage and Install plugins > QuickMapServices > Install
There are two ways of making grids:
- (Option 1) Clicking in each corner using a mosaic as reference
- (Option 2) Uploading the four-points-shapefile (e.g., GPS coordinates file or external layer shp.)
As an example let´s use the option 1: The function starts by setting four points at the field trial corners according to the steps highlighted below. The points sequence must be (1st point) left superior corner, (2nd point) right superior corner, (3rd point) right inferior corner, and (4th point) left inferior corner. At this point, the experimental borders can be eliminated (check the example). The mosaic will be presented for visualization with the North part on the superior part (top) and the south in the inferior part (bottom). The next step is provide the number of columns and rows.
Attention: The plots are identified in ascending order from left to right and bottom to top being evenly spaced and distributed inside the selected area independent of alleys.
One matrix can be used to identify the plots position according to the image above. The user can creat a matrix (manually built) informing plots ID. For instance, the new column PlotID will be the new identification. You can download an external table with field data example here: fieldData.csv and a fieldMap matrix with plots ID example here: fieldMap.csv
Check below some examples of
fieldData.csvandfieldMap.csv. Important to make sure that fieldMap is reflecting the real position of each plot in the field:
After creating the grid_shapefile right click and Open Attribute Table to check if the plots are correctly positioned:
Users can use a specific plot size to build the grid shape file by informing X and Y plot length in the parameter x_plot_size and y_plot_size. It´s very important to highlight the applied values are connected with the mosaic resolution and unit. For example, x_plot_size=0.6 means 0.6m and x_plot_size=5 means 5m or (60cm by 400cm) in the example below
Coloring and visualizing grid_fieldshape based on traits imported from fieldData.csv:
Attention: There is another great software/tutorial provided by Diego Gris called
draw-plots-qgiswith some other ways to biuld grig_shapefiles, check it at: https://github.com/diegojgris/draw-plots-qgis
- Option-01: Draw_plots_from_Excel
- Option-02: Draw_plots_from_clicks
- Option-03: Draw_plots_from_points
- Option-04: Draw_plots_from_polygons
QGIS has great tools to edit shapefiles and plots. For instance, the user needs first to activete the Advanced Digitizing Toolbar.
- Activate it at View > Toolbars > Advanced Digitizing Toolbar
- Editing plots is very simple and intuitive:
In the other hand, if you prefer editing the raster to fit better in the grid-shapefile, there is an amazing interactive georeferencing QGIS plugin called “Freehand raster georeferencer”. It allows you to (i) move, (ii) rotate, (iii) scale, (iv) adjust sides, and (v) georeference the raster using 2 points.
- Attention: Another important application for raster edition are those cases where there is more than one flight with no georeferencing or GCP (ground-control-points) during the orthomosaicing. This tool allows to make sure that all orthos/layers will align and fit well to the same plot-grid-shapefile before data extraction. Such a great tool, cheers to the authors!
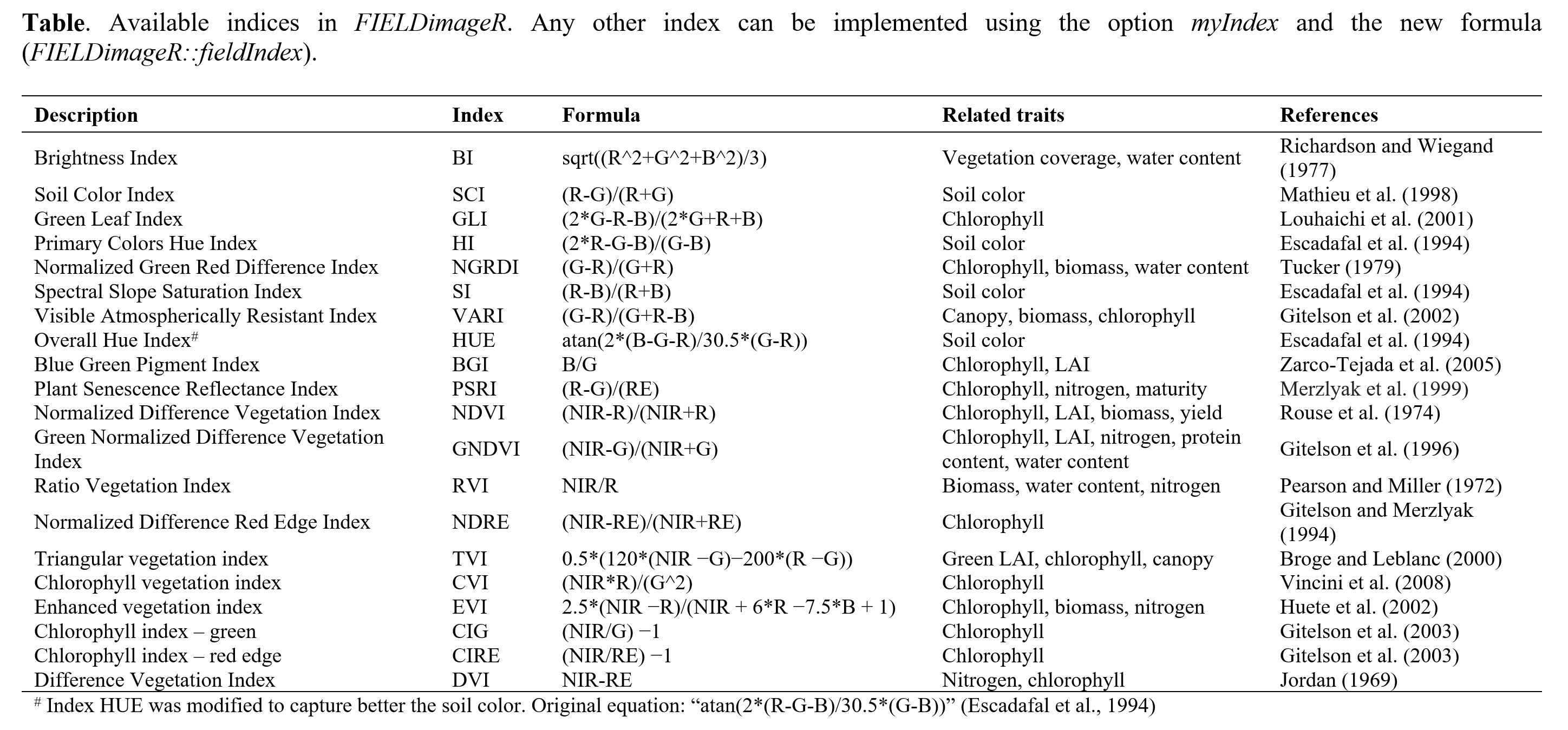
A general number of indices are implemented in FIELDimageR-QGIS using the function
fieldIndex.
Calculating vegetation indices is very simple. Users must provide the sequence of bands/wavelength avaliable in the raster. As an example, the layers must be writed 'Red,Blue,Green' for RGB. Attention: it must be 'Red' and NOT 'red'; 'Blue' and NOT 'blue'; 'Green' and NOT 'green'. Users also can calculate any other index not provided in the table above by using the box
MyIndex. Just type each index equation separated by "," (comma).
- Attention: You need to write a name as 'Index.tif' or save the output_index in a folder (Do not forget to write/save it otherwise the function doesn´t work) !!!
To better visualize specific vegetation index you can coloring it by doing a duble click at Indices layer and folowing the steps below:
Multispectral images also can be used. For instance, it is important to write the right order of layers, for example Blue,Green,Red,RE,NIR. It must be write 'RE' and NOT 'RedEdge'. You can download one example of Multispectral here: EX1_5Band.tif
Attention: In case of images/mosaics with more than 5 bands, it is important to type the desired order of layers. The names Blue,Green,Red,RE,NIR are obligatory. The vegetation indices will be calculate according to the equations described in the table above. For example, if the mosaic has 10 bands the user can type (suggestions):
-
(i) Blue,Green,Red,RE,NIR,6,7,8,9,10
-
(ii) Band1,Blue, Band 2, Green, Band 3,Red, Band 4,RE, Band 5,NIR
-
(iii) Blue,Green,Red,ban4, ban5, ban6, ban7, ban8,RE,NIR
-
(iv) Etc.
The traditional way to remove soil effect by image segmentation called Thresholding Method was implemented in the function
fieldMask. Users can choose the mask_layer_name (vegetation index from fieldIndex), the mask_crop_value, and if the cropping must be above (TRUE) or below (FALSE) the thresholding value (crop_above checkbox).
- Attention: You need to write a name as 'NewMosaic.tif' or save the output_mosaic in a folder (Do not forget to write/save it otherwise the function will not save the cropped new mosaic) !!!
Checking the quality of image segmentation to make sure the soil was removed:
The function * exact_extract* from exactextractr was adapted for agricultural field experiments through function
fieldInfo, where users can extract the mean, max, min, and sum per plot:
Checking the Attribute Table to make sure data were extracted per plot:
Visualizing extracted data and coloring plots according to the average value by doing a duble click at output shapefile info and folowing the steps below:
The plant height can be estimated by calculating the Canopy Height Model (CHM) and biomass by calculating Canopy Volume Model (CVM). This model uses the difference between the Digital Surface Model (DSM) from the soil base (before there is any sproute, Download EX_DSM0.tif) and the DSM file from the vegetative growth (once plants are grown, Download EX_DSM1.tif). To calculate CHM and CVM we used the function
fieldHeight, where CVM=cellSize(CHM)*CHM.
To better visualize the Canopy-Height-Model (CHM) and Canopy-Volume-Model (CVM) you can coloring it by doing a duble click at output CHM or output CVM and folowing the steps below:
Attention: In case the user has only one flight (e.g., one DSM) the function
fieldInterpolatecan be used to creat the soil-reference based on sampled points at the DSM. First of all, the user needs to creat a point layer shp on Layer > Creat Layer > New Shapefile Layer making sure to use the same CRS of the DSM mosaic (steps below). Now, with the functionfieldInterpolatethe user can informe the DSM (with plants) and Points.shp to creat the new Interpolated DSM to be used as soil base when calculating the Canopy-Height-Model (CHM) and Canopy-Volume-Model (CVM) withfieldHeight.
FIELDimageR-QGIS can be used to evaluate the canopy percentage per plot or LAI (Leaf Area Index). The mask output from
fieldMaskand the grig_fieldshape output fromfieldShapemust be used. Function to use:fieldArea.
To better visualize AreaPercentage or Canopy you can coloring it by doing a duble click at output fieldArea and folowing the steps below:
Checking if the values make sense:
FIELDimageR-QGIS can be used to evaluate stand count during early stages. A good weed control practice should be performed to avoid misidentification inside the plot. The mosaic_NoSoil (or output_NewMosaic) output from
fieldMaskand the grid_fieldshape output fromfieldShapemust be used. Function to use:fieldCount.
This function generates:
- New shapeFile with objects in the PLOT (Attention: total, mean, var, sd data per plot will be calculated in the grid for count, area, perimeter, and width).
- New shapeFile of single OBJECTS (Attention: The data per object is area, perimeter, width, x and y position).
The WATERSHED algorithm from EBImage-R-package can be used to identify objects by typing a tolerance value. We suggest using 0 or 1 for initial investigations.
Saving projects is very easy in QGIS. The format is
Project.qgz:
Shapefiles (e.g., grids, points, polygons, etc.) and images can be saved by selecting the layer and clicking on Export. Make sure to select the right Format, for example the
ESRI Shapelifefor grids_fieldshape. Also, the right CRS, for our example isEPSG:32616 - WGS 84:
Many times when developing algorithms it is necessary to crop the mosaic using the plot fieldShape as a reference and sort/store cropped plot images on specific folders. For instance, the function
fieldCropGridallows cropping plots and identification by 'plotID'. The user also can save each plot according to a 'classifier' logic (Attention: a column in the 'fieldShape' with the desired classification must be informed). In the example below, each plot in the 'EX1_RGB' mosaic is being cropped according to the 'grid_shape' shapefile, identified by the information in the 'plot' column, and stored/saved in specific folders with different levels of Maturity as 'classifier'.
Attention: There is NO output returned to QGIS. All cropped plots will be saved in the output_directory.
Checking cropped plots .jpg classified in each Maturity level folder:
FIELDimageR-QGIS introduce the function
fieldKmeansthat uses the K-means unsupervised method for image segmentation. This function clusters pixels on the number of clusters decided by the user. Each cluster can be associated with plants, soil, shadows, etc.
Attention: Each image/mosaic will have different cluster numbers representing plants or soil. In this example, cluster_01 is plants and cluster_02 is soil.
FIELDimageR-QGIS has one function called
fieldSegmentthat uses samples of images and machine learning algorithms for pixel classification (Supervised method). There are two models included Random Forest (default) and cart. Initially, users need to create training samples by creating a Polygon Layer (example below) or Point Layer to digitize spatial object (e.g. soil, plant, shadow etc.). For instance, users need to utilize the draw polygon or draw rectangle tool.
Selected training samples by creating polygons representing CLASS="soil" and CLASS="plant":
Checking Attribute Table IDs and CLASS columns according to the selected training samples:
After creating the training dataset, the function is very simple and intuitive to use:
Checking segmentation results based on the selected training samples:
The Output_Segment (Plants = 1 and Soil = 2) can be used to remove soil pixels by applying it as the Index_Layer input at the fieldMasck() function to crop the RGB image according to the selected criteria. In the example below, we cropped all pixels above 1.5 to eliminate the soil and select all plants. Remember to write 'Band 1' as Mask_Layer_Name:
There are many interesting ways to use the UAV data for plant breeding. The first way is evaluating single flights for specific time point (e.g., evaluating maturity, plant height, or stand count). These traits normally occur in a specific moment during the season and can be evaluated with 1 or few flights. On the other hand, traits such as plant development, diseases, biomass, and yield can be evaluated throughout the entire season using UAV data. In this case, one good strategy is combining all flights in the same analysis by calculating the area under the curve (AUC) for one trait over the time. Using AUC is an interesting way to observe and compare different biological paths and use only one general trait (e.g., growing performance, diseases resistance, etc.) for applying selection. FIELDimageR-QGIS has one function called
fieldAUCthat uses indices-raster from different time points (DAP = days after planting) to calculate the AUC per plot and save it as shp at the Output_AUC. In the example below the AUC was calculate for NGRDI index over 30, 70 and 100 DAP in potato breeding. Download the raster example here: fieldAUC_Examples.zip.
Merge two or more overlapping rasters into one new raster using
fieldMultispectral. Normally, orthomosaicing software provide single band rasters for multispectral images. This function allows to merge these single rasters in one Multispectral.tif. The same function can be used to merge other type of overlaying raster (e.g., vegetation indices). Download the raster example here: fieldMultispectral_Examples.zip.
This discussion group provides an online source of information about the FIELDimageR package. Report a bug and ask a question at:
- https://groups.google.com/forum/#!forum/fieldimager
- https://community.opendronemap.org/t/about-the-fieldimager-category/4130
Help improve FIELDimageR-QGIS pipeline. The easiest way to modify the package is by cloning the repository and making changes using R projects.
If you have questions, join the forum group at https://groups.google.com/forum/#!forum/fieldimager
Try to keep commits clean and simple
Submit a pull request with detailed changes and test results.
Let's work together and help more people (students, professors, farmers, etc) to have access to this knowledge. Thus, anyone anywhere can learn how to apply remote sensing in agriculture.
The R/FIELDimageR package as a whole is distributed under GPL-3 (GNU General Public License version 3).
-
Pawar P & Matias FI. FIELDimageR-QGIS (2024). (submitted)
-
Pawar P. & Matias FI. FIELDimageR.Extra: Advancing user experience and computational efficiency for analysis of orthomosaic from agricultural field trials. The Plant Phenome J. 2023; https://doi.org/10.1002/ppj2.20083
-
Matias FI, Caraza-Harter MV, Endelman JB. FIELDimageR: An R package to analyze orthomosaic images from agricultural field trials. The Plant Phenome J. 2020; https://doi.org/10.1002/ppj2.20005
Popat Pawar
- GitHub
- E-mail: [email protected]
Filipe Matias
- GitHub
- E-mail: [email protected]