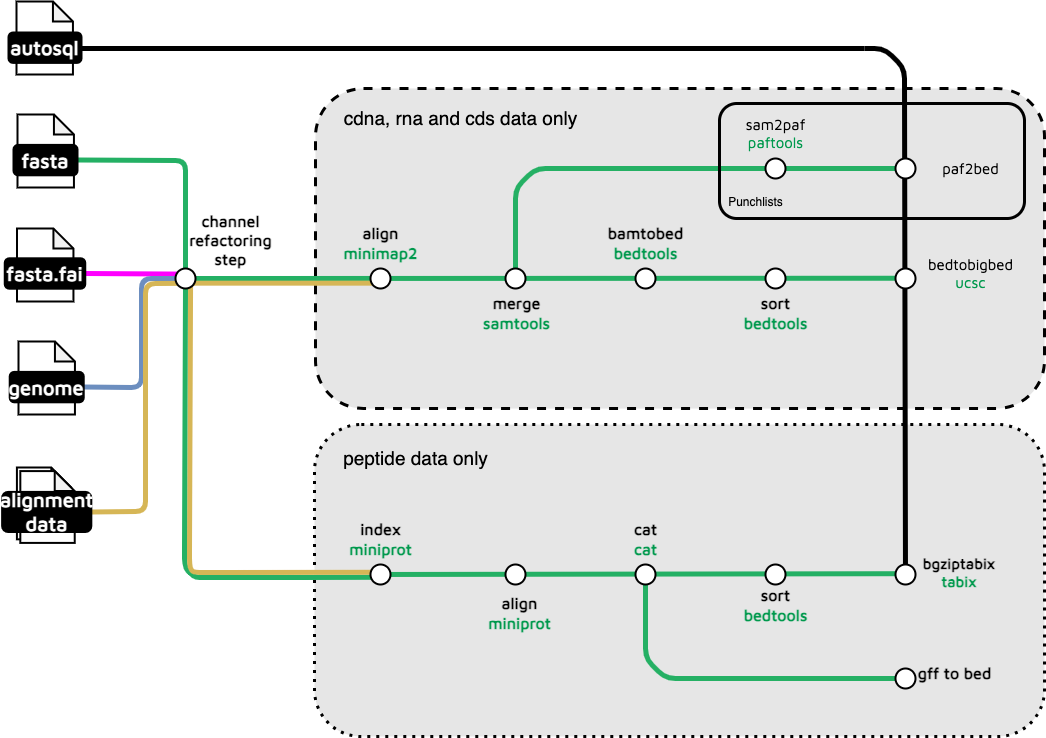
This subworkflow is used in hic_mapping as the peptide aligner.
This DDL has function calls explained below. Most of the rest of the DDL is not going to be needed other than to figure out exactly how each function gets parameters supplied to the actual command lines.
workflow PEP_ALIGNMENTS {
take:
reference_tuple // Channel: tuple [ val(meta), path(file) ]
pep_files // Channel: tuple [ val(meta), path(file) ]
max_scaff_size // Channel: tuple val(size of largest scaffold in bp)
main:
ch_versions = Channel.empty()
//
// MODULE: CREATES INDEX OF REFERENCE FILE
//
MINIPROT_INDEX ( reference_tuple )
ch_versions = ch_versions.mix( MINIPROT_INDEX.out.versions )
//
// LOGIC: GETS LIST OF META AND PEP FILES FROM GENE_ALIGNMENT
// COMBINES WITH MINIPROT_INDEX OUTPUT
// CONVERTS TO TWO TUPLES FOR PEP DATA AND REFERENCE
//
pep_files
.flatten()
.buffer( size: 2 )
.combine ( MINIPROT_INDEX.out.index )
.multiMap { pep_meta, pep_file, miniprot_meta, miniprot_index ->
pep_tuple : tuple( [ id: pep_meta.id,
type: pep_meta.type,
org: pep_meta.org
],
pep_file )
index_file : tuple( [ id: "Reference",
],
miniprot_index )
}
.set { formatted_input }
//
// MODULE: ALIGNS PEP DATA WITH REFERENCE INDEX
// EMITS GFF FILE
//
MINIPROT_ALIGN (
formatted_input.pep_tuple,
formatted_input.index_file
)
ch_versions = ch_versions.mix( MINIPROT_ALIGN.out.versions )
//
// LOGIC: GROUPS OUTPUT GFFS BASED ON QUERY ORGANISMS AND DATA TYPE (PEP)
//
MINIPROT_ALIGN.out.gff
.map { meta, file ->
tuple(
[ id : meta.org + '_pep',
type : meta.type ],
file
)
}
.groupTuple( by: [0] )
.set { grouped_tuple }
//
// MODULE: AS ABOVE OUTPUT IS BED FORMAT, IT IS MERGED PER ORGANISM + TYPE
//
CAT_CAT (
grouped_tuple
)
ch_versions = ch_versions.mix( CAT_CAT.out.versions )
//
// MODULE: SORTS ABOVE OUTPUT AND RETAINS GFF SUFFIX
// EMITS A MERGED GFF FILE
//
BEDTOOLS_SORT (
CAT_CAT.out.file_out ,
[]
)
ch_versions = ch_versions.mix( BEDTOOLS_SORT.out.versions )
//
// MODULE: CUTS GFF INTO PUNCHLIST
//
EXTRACT_COV_IDEN (
CAT_CAT.out.file_out
)
ch_versions = ch_versions.mix( EXTRACT_COV_IDEN.out.versions )
BEDTOOLS_SORT.out.sorted
.combine( max_scaff_size )
.map {meta, row, scaff ->
tuple(
[ id : meta.id,
max_scaff : scaff >= 500000000 ? 'csi': '' ],
file( row )
)
}
.set { modified_bed_ch }
//
// MODULE: COMPRESS AND INDEX MERGED.GFF
// EMITS A TBI FILE
//
TABIX_BGZIPTABIX (
modified_bed_ch
)
ch_versions = ch_versions.mix( TABIX_BGZIPTABIX.out.versions )
MINIPROT_INDEX uses conda "bioconda::miniprot=0.11=he4a0461_2" and calls
miniprot \\
-t $task.cpus \\
-d ${fasta.baseName}.mpi \\
$args \\
$fasta
miniprot_align runs
miniprot \\
$args \\
-t $task.cpus \\
${ref} \\
${pep} \\
> ${prefix}.${extension}
CAT_CAT is also used in kmer and uses "conda-forge::pigz=2.3.4" and the call depends on the extensions:
// | input | output | command1 | command2 |
// |-----------|------------|----------|----------|
// | gzipped | gzipped | cat | |
// | ungzipped | ungzipped | cat | |
// | gzipped | ungzipped | zcat | |
// | ungzipped | gzipped | cat | pigz |
prefix = task.ext.prefix ?: "${meta.id}${file_list[0].substring(file_list[0].lastIndexOf('.'))}"
out_zip = prefix.endsWith('.gz')
in_zip = file_list[0].endsWith('.gz')
command1 = (in_zip && !out_zip) ? 'zcat' : 'cat'
command2 = (!in_zip && out_zip) ? "| pigz -c -p $task.cpus $args2" : ''
"""
$command1 \\
$args \\
${file_list.join(' ')} \\
$command2 \\
> ${prefix}
BEDTOOLS_SORT uses conda "bioconda::bedtools=2.31.0" to run
bedtools \\
sort \\
-i $intervals \\
$genome_cmd \\
$args \\
> ${prefix}.${extension}
EXTRACT_COV_IDEN has a /tree/bin shell script called as
extract_cov_iden.sh ${file} ${prefix}.bed
That script is:
#!/bin/bash
# extract_cov_iden.sh
# -------------------
# A shell script to convert a
# extract coverage information from
# PAF header and reorganising the data
# -------------------
# Author = yy5
version='1.0.0'
if [ $1 == '-v'];
then
echo "$version"
else
grep '##PAF' $1 | sed 's/##PAF\t//g'|awk 'BEGIN{FS="\t";}{a[$1]++;if(a[$1]==2)print v[$1] ORS $0;if(a[$1]>2)print;v[$1]=$0;}' | awk '$(NF+1) = ($10/$11)*100'|awk '$(NF+1) = ($10/($2*3))*100'|awk -vOFS='\t' '{print $6,$8,$9,$1,$2,$10,$(NF-1),$NF}' > $2
fi
Another bedtools sort and a tabix step complete the subworkflow.