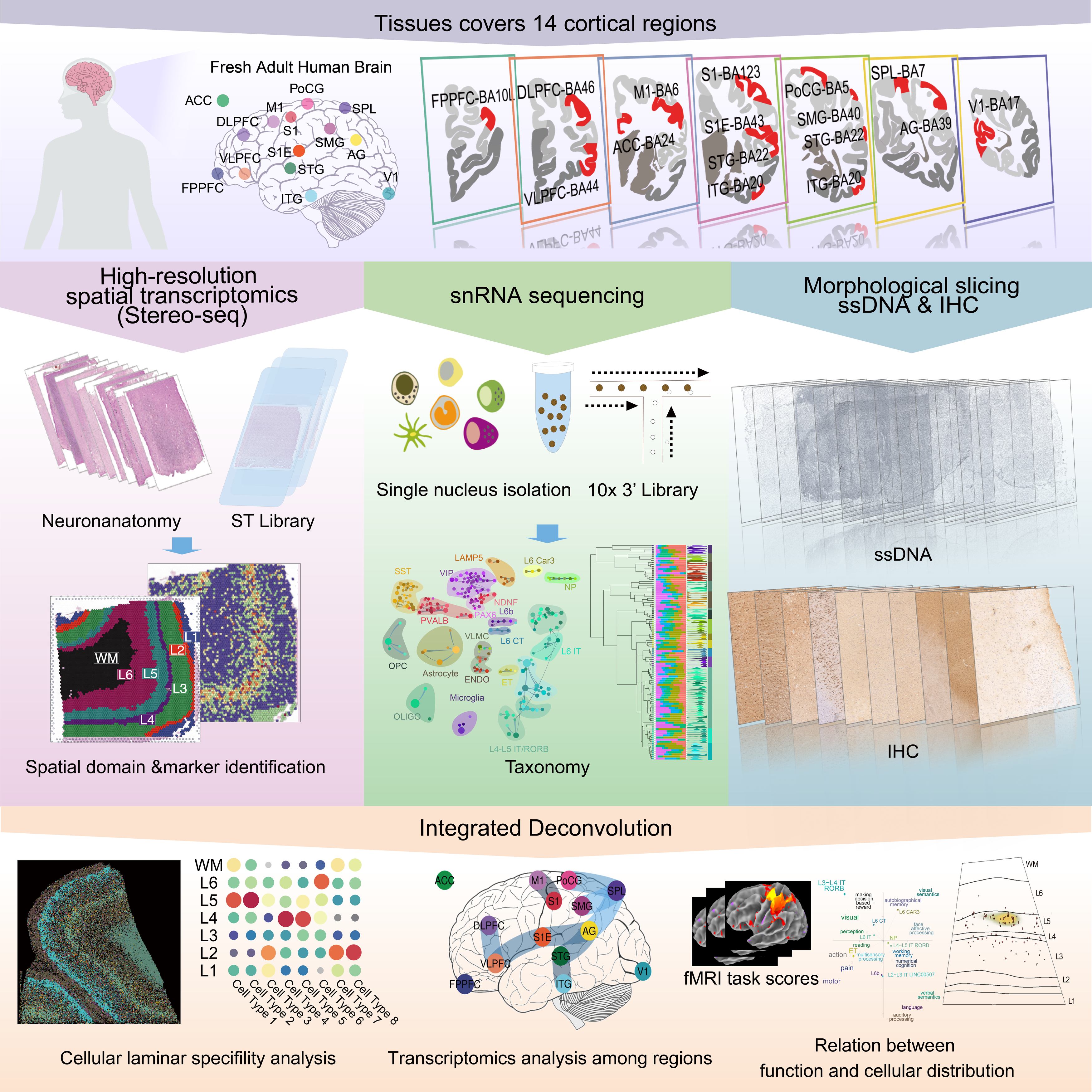
This project includes the complete workflow for single-cell and Stereo-seq spatial transcriptomics analysis, covering data quality control, integration analysis, spatial domain annotation, and main figure generation. The workflow is divided into three sections:
- Download specific cellxgene BICCN WHB single-cell data.
- Perform quality control on the raw single-cell data from this study.
- Integrate the processed single-cell data with BICCN datasets.
- SquareBin Quality Control: Conduct basic quality control on Stereo-seq data.
- CellBin Quality Control: Further process the CellBin data.
- Spatial Domain Clustering and Annotation: Perform clustering and annotation of spatial domains based on SquareBin data.
- RCTD Deconvolution Annotation: Use single-cell data and RCTD to annotate spatial domains via deconvolution.
The figure generation scripts are organized according to the order of the manuscript's figures. Run the corresponding scripts after completing the single-cell and Stereo-seq analyses to generate the final results.
Please follow the workflow in sequence to ensure data processing consistency. For any questions, contact the project lead.