The source code for paper published in "Progress in Artificial Intelligence" journal. DOI: 10.1007/s13748-024-00314-3.
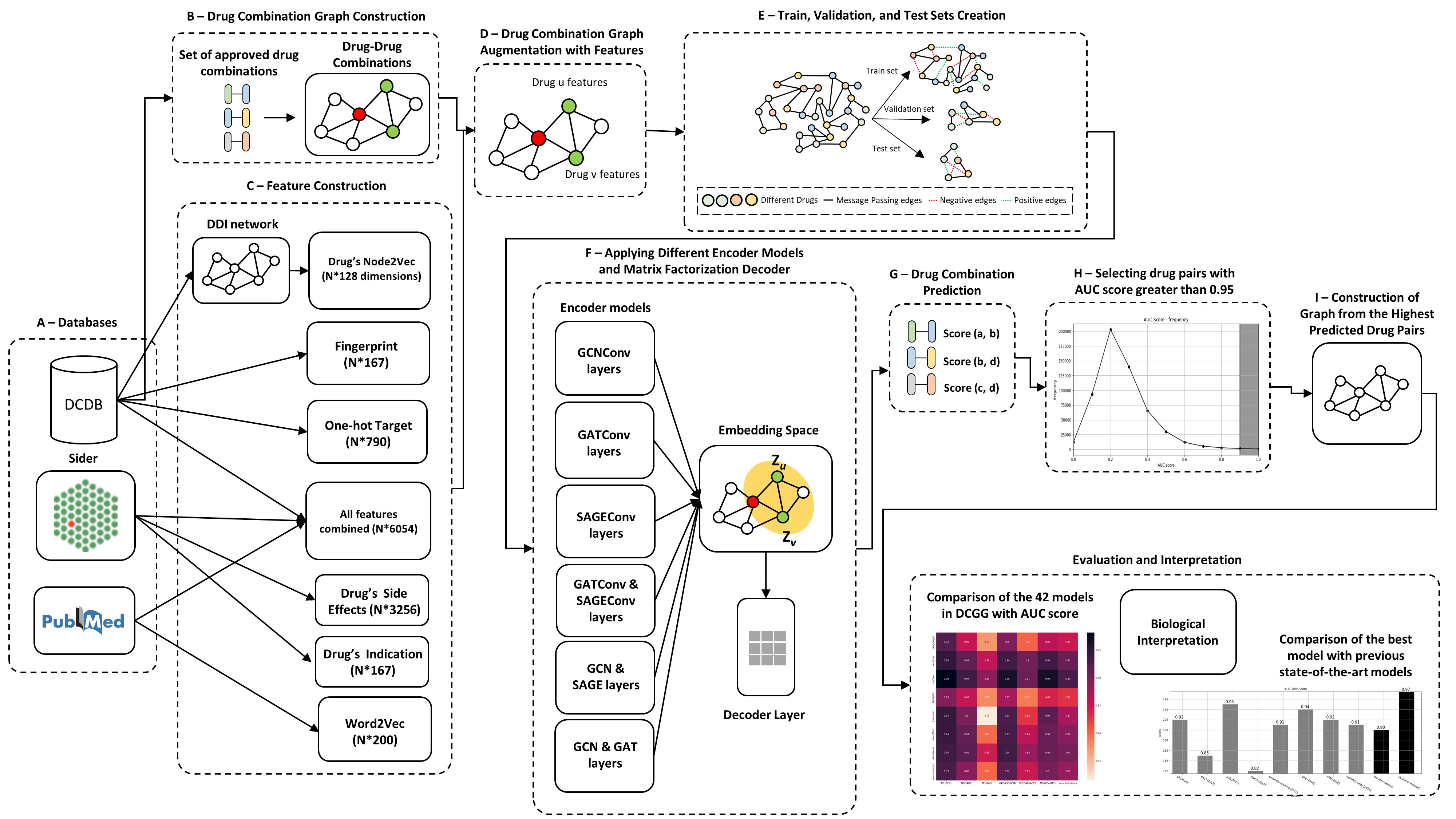
DCGG is a novel approach towards drug combination prediction utilizing graph neural networks and graph auto encoders. The approach comprises of 6 different models and 7 different graphs. The graphs are augmented with differenet node features including drug indicatinos, drug side-effects, and node2vec drug features extracted from drug-drug-interaction network.
Submitted to Briefings in Bioinformatics
Authors: S.S. Ziaee, H. Rahmani, M. Tabatabaei, Anna H.C. Vlot, Andreas Bender
DCGG main steps:
Notes:
To run the codes please use these versions of softwares and libraries:
Python 3.9, Pytorch 1.11, torch geometric 2.0.4, Numpy 1.18.1, and Pandas 1.0.1.