-
Notifications
You must be signed in to change notification settings - Fork 24
Apache_Spark_and_VisIt
Apache Spark is, IMHO, an improved implementation of Hadoop Map-Reduce, a basic technology used in Big Data Science in extreme scale commercial computing.
Superficially, commercial Big Data computing and DOE HPC computing appear to be vastly different paradigms of extreme scale computing. The former is optimized to process large volumes of tiny pieces of unstructured text data while the latter is optimized to process small volumes of extremely large structured (not in a mesh topology sense but in a well-defined organization of related data sense) meshes and fields over meshes.
At LLNL, we won approval for a 2 week, Idea Days project to investigate the use of Big Data tools for some common HPC visualization and post-processing operations and compare software engineering aspects of the two approaches. In the past, we’ve looked at the Silo, Hadoop and VisIt. In this case, we decided to look at Apache Spark. Apache Spark supports Java, Scala and Python programming language scripting interfaces. We developed capabilities here using Apache Spark’s Python interface, py-spark.
The first question we considered is how to export HPC data (e.g. contents of Silo, Exodus, Xdmf, etc. data files) to a suitable Big Data organization.
A basic question is how to design a single, coherent Spark-oriented database capable of storing scientific computing mesh and field data in a way that achieves several goals
- Is designed to help shed light on longer term applicability of Big Data approaches to post-processing and analysis tool development in HPC scientific workflows
- In other words, we aren’t aiming to get something, anything, working solely for the purposes of completing just this tiny project. We want the work to help identify issues and options if we were to wholly replace current practices with this new, Big Data approach.
- Stores scientific computing mesh and field data that is consistent with text-oriented, key-value pair processing for which Big Data science software stacks are optimized
- This has the added benefit of not requiring any customization of the Java-based interfaces to persistent storage used within the Big Data stack
- Keys in the input database identify individual mesh elements (e.g. zones and the nodes that comprise them), uniquely over the entire input database
- Given a node key, for example, it is possible to determine which domain from among many domains of the mesh, which mesh from among many meshes of a given timestep, which timestep from among many timesteps of a given database, which database from among many databases of a given user and which user from among many users of an entire computing center’s user base.
- In addition, a mesh entity key (node or zone) is designed to also indicate the topological dimension of the entity (0,1,2 or 3) as well as whether the entity is a ghost entity or not
- Enables variable scoped queries over the resulting database
- The narrowest scope being a single block of a single mesh of a single timestep of a single database of a single user
- The widest scope being all blocks of all meshes of all timesteps of all databases of all users
- Variations in query scope between these extremes must be easily supported
- Compute resources (processor count and/or memory size) necessary to satisfy any given query on the database are determined by query scope and not by overall input database size
- The input database design makes it possible to read from persistent storage only the data needed to satisfy a given query
- Avoid a design that forces applications to read everything about a mesh in order to query anything about a mesh
- Optimizes persistent storage efficiency to the maximum extent possible within constraints of other design goals specified here
The most pressing question in the design is the construction of mesh entity keys. A key needs to uniquely identify a mesh node or zone over all blocks, meshes, timesteps, databases and users. To simplify things, we first divide a mesh entity key into sub-fields for each of these classes of things. Next, by selecting appropriate limits on the maximum possible number of things we will permit in each class, we can determine the number of digits needed to represent each sub-field of the key and then the overall key.
To unambiguously identify one object from among N objects, we need a key with log B (N) digits using a number system with 0…B-1 digit characters. In the database, we will be storing these keys as ASCII strings. So, we have some flexibility over the choice of digit characters. If we were to use just binary digit characters (only the characters 0 or 1_), then we would wind up using 8 bits (an ASCII character is 8 bits) for each digit character representing only a single bit of precision in the key. That represents an 8x storage inefficiency. If we use just the decimal characters (_0 … 9), the storage inefficiency is 8 / log 2 (10) or 2.4x, which is still quite large. So, we choose to use a base 64 ASCII digit character set to represent mesh entity keys. The unique digits in this 64-character set are 0-9a-zA-Z%#. The smallest digit character is 0 and the largest is ‘#’ (representing the value 63 in base 10). With this approach, each 8-bit ASCII digit character in a mesh entity key represents 6 bits of actual precision and the storage inefficiency is only 25%.
Next, we set suitable upper limits on the total number of possible things in each class for each sub-field of the key and we arrive at the following mesh entity key design.
| Field | User Id | Database Id | State Id | Mesh Id | Block Id | Entity Id | Total # of Digit Chars | #Bits used | #Bits storage |
|---|---|---|---|---|---|---|---|---|---|
| Limit | 4096 | 262,144 | 4096 | 64 | 16,777,216 | 68,719,476,736 | |||
| B64 Digits | 2 | 3 | 2 | 1 | 4 | 6 | 18 | 108 | 144 |
| B10 Digits | 4 | 6 | 4 | 2 | 8 | 11 | 35 | 280 |
In the table above, we indicate the number of digits needed in each sub-field for both our base-64 number system and a base-10 system. Note that the base-10 approach would require nearly twice the number of ASCII digits in each key, reducing storage efficiency by 2×. On the other hand, an advantage of a base-10 approach is that the resulting numbers are human readable. However, without the inclusion of special field separator characters (which would increase the key size by 5 characters) to break the key into its sub-fields, even a base-10 coded key is going to be difficult for humans to decode zone or node numbers, block numbers or time steps.
From the above table, the design we’ve taken here can support a maximum of 4096 users. Each user can, over their lifetime, produce a maximum of 262,144 databases. Note that a database here is not the contents of a single Silo or netCDF or Xdmf file but the contents of a whole ensemble of such files representing different states of the same simulated problem. Each such database can contain a maximum of 4096 states. Each state can contain a maximum of 64 different mesh objects and each mesh can contain a maximum of almost 17 million blocks. Finally, any single block can contain a maximum of almost 69 Trillion mesh entities (nodes and zones). It is believed that these limits are in the ballpark of the needs for LLNL’s computing center.
Why do we need to support almost 69 Trillion mesh entities on any single block? I mean, in a large block-decomposed mesh, we can have a lot of mesh entities. But they can be uniquely identified either by a single, very large range global numbering or a pair, {block, local entity}, of much smaller range numberings. Here allowing for 69 Trillion possibilities in the mesh entity subfield when we already have a subfield in the key that disambiguates them by block seems like overkill. We are doing this for a few reasons. First, we do need to allow for those unusual cases where we have very, very large monolithic (e.g. non-decomposed, typically structured) meshes such as are common in parallel-distributed arrays approaches in HPC simulations. Another reason is that with a base-64 numbering, the jump in upper limits between a few character variation in the subfield width is so large, we opted to play it on the safe side at the expense of maybe one or two additional characters in the key. Finally, we also want to encode into the key some additional information; the topological dimension of the mesh entity and whether the mesh entity is a ghost entity or not.
So, the last bit of our key design involves the following constraints on the Entity Id subfield. Given the nth mesh entity in a block, we encode the Entity Id subfield as 8n+{0,1,2,3} if it is a point(0), edge(1), face(2) or volume(3) element and 8n+{4,5,6,7} if it is a ghost point, edge, face or volume entity. So, given a mesh entity key in our database, if the remainder after dividing by 8 is say 3, we know it is a non-ghost volumetric entity. If it is 5, we know it is a ghost edge entity.
Note that in native HPC applications, mesh entity keys in C/C++ and Fortran codes are routinely implemented as 4- or 8-byte integer indices. So, our 18 character keys here are about 2-4x less efficient in storage. On the other hand, the keys we construct here have the potential to remain unique across a much wider scope.
Ok, now that we’ve decided how to identify mesh entities within this large, all-encompassing textual database, the next question is how we lay mesh and field data out in files and then files in a directory hierarchy.
Finally, Eric Brugger mentioned it would be more efficient as well as more accurate to also capture the floating point data using the same method to encode binary data as ascii.
After designing this encoding, I’ve since learned that this kind of issue, encoding arbitrary numerical data into ascii digits, is a common problem to which there are many solutions. It is known as a Base64 encoding. In fact, Python has builtin support for a number of different Base64 encodings. A future implementation would most likely be better off simply using one of the available encodings. Like many other Base64 encodings, I used the same 62 initial characters. I tried to avoid characters that might have meaning to the shell or in regular expressions.
Part of the approach we take here is inspired by the work of Cyrus Harrison and Kathleen Biagas on a new data model for VisIt’s in-situ interface. As such, the focus here is on the representation of an individual block of mesh. Here, our concern is representing a mesh block in terms of one or more text files. Our approach is simple and straightforward. We store the following key-value text files…
- coords.txt
- Keyed by node entity keys
- Value is zero or more columns of Cartesian coordinate tuples
- A set of node keys with no coordinates is possible for graph-like data where specific spatial locations are often not needed and only the relationships (edges) in the graph are important.
- Support for alternative coordinate systems such as Spherical or Cylindrical will require minor modifications
- Example (2nd column indicates ghost status, remaining columns for 3D spatial data)
0000n004000h0001aM,0,0.384448,4.88489,8.0
0000n004000h0001aU,0,0.392293,4.98458,8.0
0000n004000h0001b4,1,-1.56522e-06,3.9,8.0
.
.
.
- .txt
- Keyed by node entity keys
- Value is one or more components values
- Scalar variables 1 component, vector variables 2 (2D), 3 (3D), symmetric tensor 3 (2D), 6 (3D), full tensors, 4 (2D), 9 (3D), and array variables otherwise
- Repeated for each nodal variable where is the name of the variable
- Example (scalar variable)
0000n004000h0001aM,0.204078
0000n004000h0001aU,0.199996
0000n004000h0001b4,0.256404
.
.
.
- topology.txt
- Always present even for structured meshes
- May not be needed for meshes consisting solely of nodes
- Optimizing this away for structured grids will require customization of HDFS InputFile classes
- Keyed by mesh zone entity keys
- Value is mesh entity type, number of nodal entities and then the nodal entity keys
- For zonal mesh entities specified in terms of faces and then either edges or nodes, will require some modification
- Example (several hex elements; 2nd col is ghost status, 3rd is element type, 4th is #nodes, remaining is node keys)
0000n004000h0000LX,0,12,8,0000n004000h0000UM,0000n004000h0000Tg,0000n004000h0001aM,0000n004000h0001cg,0000n004000h0000UE,0000n004000h0000T8,0000n004000h0001aE,0000n004000h0001c8
0000n004000h0000M3,0,12,8,0000n004000h0000UY,0000n004000h0000Ts,0000n004000h0001aY,0000n004000h0001cs,0000n004000h0000UQ,0000n004000h0000Tk,0000n004000h0001aQ,0000n004000h0001ck
0000n004000h0000Mb,0,12,8,0000n004000h0002QQ,0000n004000h0002Pk,0000n004000h00036Q,0000n004000h00038k,0000n004000h0002QI,0000n004000h0002Pc,0000n004000h00036I,0000n004000h00038c
.
.
.
- Always present even for structured meshes
- .txt
- Keyed by zone entity keys
- Value is one or more component values
- Scalar variables 1 component, vector variables 2 (2D), 3 (3D), symmetric tensor 3 (2D), 6 (3D), full tensors, 4 (2D), 9 (3D), and array variables otherwise
- Repeated for each zonal variable where is the name of the variable
- Example (scalar variable)
0000n004000h000003,0.028
0000n004000h00000b,0.028
0000n004000h00000j,0.028
.
.
.
- materials.txt
- Present only when the mesh has materials
- Keyed by zone entity keys
- Value is fractional volume each material occupies in the zone
- This is hugely inefficient, particularly for clean material problems
- Improvement in storage efficiency here is unclear
- Example (3 material problem, zones here are clean in material 0)
0000n004000h0000LX,1,0,0
0000n004000h0000M3,1,0,0
0000n004000h0000Mb,1,0,0
.
.
.
First, there is a lot of miscellaneous metadata and some key mesh and field data that this approach does not easily support. Some of this un-captured data is probably fairly easy to deal with with minor modifications to our approach. Some may require more significant study.
- Minor design modifications to support
- Coordinate systems other than Cartesian
- Domain naming conventions
- Block naming conventions
- Material names
- Material species names
- Units on variables
- Labels on variables
- Component names
- Missing values
- Intensive/Extensive quantities
- More significant design modifications to support
- Block-Domain relationships
- Optimizations or rectilinear coordinates and structured mesh topology
- Arbitrary subsets
- Variables defined on subsets
- Mixed material variable values
- Material Species
- Coordinate system for the state space
- This is a lot of small files solution instead of a few large files solution
- We have different files for different mesh blocks and different variables on a mesh block. This is substantially different from how data is stored in Silo files. Many mesh blocks go into one file and all the variables for a mesh block go into one file. But, the Silo interface permits us to read just some of the objects in a file without reading the whole file.
- In all likelihood, this represents a serious impedance mismatch with the low-level HDFS interface and filesystem. However, with the default I/O interfaces in HDFS, we must always read an entire file if we want to read any of it. This suggests that an implementation of this appraoch for HPC computing demains that we customize the low-level Java classes responsible for I/O in HDFS so that we can read data from HPC databases more intelligently.
Second, the approach has a number of storage inefficiencies. Some of these have already been mentioned above. To summarize, the storage inefficiencies are…
- The topology of structured meshes winds up being explicitly stored
- For a structured mesh in 2D (e.g. all quads), the additional storage cost for explicit topology is equivalent to 4 scalar fields. In 3D (all hexes), its equivalent to 8 scalar fields.
- If along with the mesh, there are 20 scalar fields, the additional 4 or 8 fields represents a 20-40% storage increase. On the other hand, if along with the mesh, there are 100 scalar fields, the storage increase is just 4-8%.
- However, these observations apply only to persistent storage where the mesh and all of its scalar fields must be stored. Typically, only the mesh plus a handful of scalar fields (or handful of components of a composite field like a 9-component Tensor valued field) is read into memory of an executing data analysis tool for any given algorithm. In this case, the storage burden for explicit topology of a structured mesh represents as much as a 4-8x storage increase.
- Mesh entity keys are 2-4x bigger
- This has the effect of increasing the storage cost of explicit topology because it is the topology data in which mesh entity keys are stored (e.g. a hexahedral zone is identified by an enumeration of its 8 node Ids). So, this winds up exacerbating storage inefficiencies described in item 1.
- On the other hand, the mesh entity keys we use here are unique over literally everything.
- Mesh entity keys are repeated in each text file holding mesh data
- In the coords file and every nodal variable file, the first column is a node key. Likewise, in the topology file, the materials file and every zonal variable file, the first column is a zone key.
- Turns out this is easily corrected by NOT storing the keys as the first column in the files to begin with but instead, storing the keys as a separate file, “nodeKeys.txt.bz2” and “zoneKeys.txt.bz2” and then immediately using Spark’s
zipmethod to combine two RDDs
- Materials are represented as dense volume fractions
- The fact that we use volume fractions at all is due the the possibility of having mixing materials. But, not all applications support or need mixing of materials.
- Worse, even when mixing materials are needed, not all zones in a mesh are mixing nor are they mixing all of the possible materials in the mesh.
- This is a hugely inefficient means of storing material data.
- Since doing this work, there are some obvious improvements we could make here. There is no requirement that each line in materials.txt contain the same number of columns (or comma separated values). For clean zones, we can just store a single integer, the material id. For mixing zones, we can store a vector of (material id, volume fraction) pairs.
- All data is stored as ASCII
- We’ve already discussed this in the context of our entity keys. The encoding we developed represents a 25% storage inefficiency. We’re storing 8-bit ASCII characters for each 6-bit (base-64 digit) in our mesh entity keys.
- For all other numerical data, the inefficiency is 2.4x, 8 / (log 2 (10)).
- Further storage inefficiencies when reading into Python
- The preceding items describe in detail the storage inefficiencies of the data as it is stored in files persistent storage.
- However, this data is going to be read into Python py-spark Scripts and then operated upon there.
- It comes into Python as String data. The smallest String object in Python is ~200 bytes. If we’re smart, as soon as possible after loading the data into Python, we’ll convert it from its original String representation to integer or floating point numbers. But an int or float in Python is still ~20 bytes. This is just something to keep in mind when developing py-spark scripts.
Nonetheless, in spite of all these inefficiencies these text files compress very well with bzip2 compression. And, Apache Spark is designed to read bzip2-compressed files natively. 7zip may provide even better compression and there appears to be plans in Apache Spark to also support 7zip files. In addition, it’s conceivable that a custom compression scheme that is designed apriori to handle only numerical data plus our pseudo-numerical entity keys could achieve even better compression.
As far as organization of data into files in a directory hierarchy, we considered primarily the use of Spark over Lustre and not necessarily anything specific to the HDFS file system. The above describes how all of the data for a single mesh block is stored. These files represent the leaves of a deep directory hierarchy organized with users at the top, then databases, then states, then meshes, then blocks and finally the individual files for each block. This directory hierarchy is structured as follows. . .

Not shown in the diagram above, at each level in the directory hierarchy, there is an index or mapping file that stores the names of all the objects at that level keyed by their integer index. For example, at the top level, the users.txt.bz2 file stores the names of all the users and their integer ids within the database. Or, for userA’s database named dbA, the file states.txt.bz2 stores all the names of the states for that database. In the current implementation, the states are named with the same 6-character State Id subfield string for mesh entities keys in that state. But, this could easily be generalized to support arbitrarily named states. Likewise for mesh blocks.
We developed a few Python scripts that use VisIt together with VisIt’s Python Queries to export any database VisIt is capable of reading into this all-encompassing Spark database.
- Python Query Script
- Driver Script
- Utilities used by the above scripts.
- VisIt HDFS database plugin to read data from the database the above tools help to construct.
To use these scripts to export a given VisIt database to the Spark friendly format described above, you would run VisIt from a directory containing the above 3 python files
<path-to-visit-home>/bin/visit -cli -nowin -s hdfs_export_driver.py <db-name> <hdfs-root-name> [<optional-key-base>]
For example, here is what exporting the multi_ucd3d.silo file looks like. . .
% ls hdfs_export.vpq hdfs_export_driver.py hdfs_export_utils.py hdfs_export_utils.pyc spark_examples % ~/visit/trunk/build/bin/visit -nowin -cli -s ./hdfs_export_driver.py ~/visit/trunk/data/silo_hdf5_test_data/multi_ucd3d.silo dbroot Running: cli -dv -nowin -s ./hdfs_export_driver.py /Users/miller86/visit/trunk/data/silo_hdf5_test_data/multi_ucd3d.silo dbroot [Exporting curve line for time index 0] VisIt: Message - [SciDB Export Complete] [Exporting curve log for time index 0] VisIt: Message - [SciDB Export Complete] [Exporting curve wave for time index 0] VisIt: Message - [SciDB Export Complete] [Exporting mesh mesh1 for time index 0] VisIt: Message - [SciDB Export Complete] [Exporting mesh mesh1_back for time index 0] VisIt: Message - [SciDB Export Complete] [Exporting mesh mesh1_dup for time index 0] VisIt: Message - [SciDB Export Complete] [Exporting mesh mesh1_front for time index 0] VisIt: Message - [SciDB Export Complete] % ls dbroot hdfs_export_driver.py hdfs_export_utils.pyc visitlog.py hdfs_export.vpq hdfs_export_utils.py spark_examples % ls -R dbroot miller86 users.txt dbroot/miller86: dbs.txt multi_ucd3d dbroot/miller86/multi_ucd3d: 000000 states.txt dbroot/miller86/multi_ucd3d/000000: line log mesh1 mesh1_back mesh1_dup mesh1_front meshes.txt wave dbroot/miller86/multi_ucd3d/000000/line: 000000 blocks.txt dbroot/miller86/multi_ucd3d/000000/line/000000: coords.txt.bz2 topology.txt.bz2 dbroot/miller86/multi_ucd3d/000000/log: 000000 blocks.txt dbroot/miller86/multi_ucd3d/000000/log/000000: coords.txt.bz2 topology.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1: 000000 000005 000010 000015 000020 000025 000030 000035 000001 000006 000011 000016 000021 000026 000031 blocks.txt 000002 000007 000012 000017 000022 000027 000032 000003 000008 000013 000018 000023 000028 000033 000004 000009 000014 000019 000024 000029 000034 dbroot/miller86/multi_ucd3d/000000/mesh1/000000: coords.txt.bz2 hist.txt.bz2 p.txt.bz2 v.txt.bz2 d.txt.bz2 m1vf_on_mats_1.txt.bz2 topology.txt.bz2 w.txt.bz2 d_on_mats_1_3.txt.bz2 materials.txt.bz2 u.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1/000001: coords.txt.bz2 hist.txt.bz2 materials.txt.bz2 topology.txt.bz2 w.txt.bz2 d.txt.bz2 m1vf_on_mats_1.txt.bz2 p.txt.bz2 u.txt.bz2 d_on_mats_1_3.txt.bz2 m2vf_on_mats_2.txt.bz2 p_on_mats_2.txt.bz2 v.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1/000002: coords.txt.bz2 hist.txt.bz2 p.txt.bz2 v.txt.bz2 d.txt.bz2 m1vf_on_mats_1.txt.bz2 topology.txt.bz2 w.txt.bz2 d_on_mats_1_3.txt.bz2 materials.txt.bz2 u.txt.bz2 . . . dbroot/miller86/multi_ucd3d/000000/mesh1/000034: coords.txt.bz2 hist.txt.bz2 materials.txt.bz2 topology.txt.bz2 w.txt.bz2 d.txt.bz2 m1vf_on_mats_1.txt.bz2 p.txt.bz2 u.txt.bz2 d_on_mats_1_3.txt.bz2 m2vf_on_mats_2.txt.bz2 p_on_mats_2.txt.bz2 v.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1/000035: coords.txt.bz2 hist.txt.bz2 p.txt.bz2 v.txt.bz2 d.txt.bz2 m1vf_on_mats_1.txt.bz2 topology.txt.bz2 w.txt.bz2 d_on_mats_1_3.txt.bz2 materials.txt.bz2 u.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1_back: 000000 000003 000006 000009 000012 000015 blocks.txt 000001 000004 000007 000010 000013 000016 000002 000005 000008 000011 000014 000017 dbroot/miller86/multi_ucd3d/000000/mesh1_back/000000: coords.txt.bz2 topology.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1_back/000001: coords.txt.bz2 topology.txt.bz2 . . . dbroot/miller86/multi_ucd3d/000000/mesh1_front/000016: coords.txt.bz2 topology.txt.bz2 dbroot/miller86/multi_ucd3d/000000/mesh1_front/000017: coords.txt.bz2 topology.txt.bz2 dbroot/miller86/multi_ucd3d/000000/wave: 000000 blocks.txt dbroot/miller86/multi_ucd3d/000000/wave/000000: coords.txt.bz2 topology.txt.bz2
The driver script is designed to be re-run multipe times each for a different input database and to insert the resulting data and files into an existing, growing database directory tree as described above. We used these escripts and exported the entire VisIt test suite data into this format. In addition, we wrote a VisIt plugin (the “HDFS” plugin) that is able to read data in this format back into VisIt. Currently, the HDFS plugin reads only the mesh. It does not read any variables on the mesh. The HDFS plugin helped to identify and correct some initial bugs in the export process. For example, in the pictures below, we illustrate a buggy Spark database on the left and the original data on the right. The VisIt plugin helped to confirm that indeed there was a bug in the export tools and later confirm we corrected it.


Exporting the entire VisIt test suite consisting of about 363 databases took approximately 3 hours on a single processor of surface.llnl.gov. In some cases, the exported data in the new format required less storage. In other cases, the exported data required more storage than the original. However, the increased storage was generally no more than about 2×. On the other hand, it is important to keep in mind that in some cases, not all of the original data is exported to the new format either. Nonetheless, in spite of several glaring inefficiencies (2-4x larger keys, keys being repeated in every variable file, explicit topology, dense material fractions, etc.) this initial test suggests that with compression these storage inefficiencies can be kept fairly low. This is much more encouraging than the more naive approach we took in a previous Silo_Hadoop_and_VisIt. In addition, in the section immediately below we mention some options to reduce inefficiencies even further or perhaps eliminate them entirely. At the same time, compression impacts only the data as it is stored in persistent storage and not the in-memory storage of the resulting data after it is read from disk. This latter issue is still a potentially serious concern that may need to be addressed.
With some customization of Java classes used internally by HDFS software stack upon which Spark is implemented (note: In commercial industry it appears quite common to specialize pieces of HDFS Java implementation to support workflows), it is conceivable that data translation we perform here could be done on the fly, as part of the process of reading data into Spark workflows. Conceivably we could leave data in Silo, netCDF, Xdmf files (or in whatever HPC format it originates), and have Java plugins that read from those native formats and then do JIT key-value pair construction described here on the fly as these files are being read into Spark. More interestingly, if the JIT key-value constructing algorithms could be given a characterization of the scope of the query (is it over one database or several, over one timestep or several, etc.), the keys could be optimized to be constructed to suit the given scope. Smaller scopes would require shorter keys. For example, if the scope is over data for only one user and one database, the User-Id and Database-Id sub-fields could be completely eliminated from the process. This would have the added benefit of saving in-memory representation in the Spark workflows.
On the other hand, given the myriad of different HPC data formats (VisIt currently supports over 130 different format readers) as well as the variations in how these formats support timesteps, blocks and multiple meshes, it would still be essential to design an over-arching abstraction similar to the File and Directory Layout described here, that would permit Spark workflows to uniformly manipulate data in these terms. This represents a volume of code that would be on the order of VisIt’s Generic Database class and everything that class uses (File Format, File Format Interfaces and all the reader plugins) to read data; about ~500,000 lines of C/C++ code. In addition, there would have to be JNI’s (Java Native Interfaces) for many of the HPC data format libraries such as Silo, netCDF, Exodus, HDF5, etc.
There appears to have already been some work done in trying to read HDF5 files’ datasets as RDDs
In retrospect, there may have been better choices for the input database design we finalized upon here. However, given the limited time available to consider options, we arrived upon the design described here.
A final comment is that these questions of how to encode data into textual form suitable for Big Data analysis is somewhat unique to the application of such techniques to HPC data. Typically, Big Data applications have little or no control over how the textual data they process may be keyed at input. They start from textual data to begin with and are not faced with the task of deciding how best to represent input data in a textual, key-value form. In all likelihood, this issue probably requires significantly more consideration than we were able to give it here.
We would now like to use the above database to explore the space of post-processing and data analysis operators as Apache Spark python scripts. Our rationale is that a majority of the code in post-processing, visualization and data analysis tools is related to the data processing aspects of the workflow. This was true even as far back as the late 1990s. A comparison of some commercial and DOE developed tools at the time, shown below,
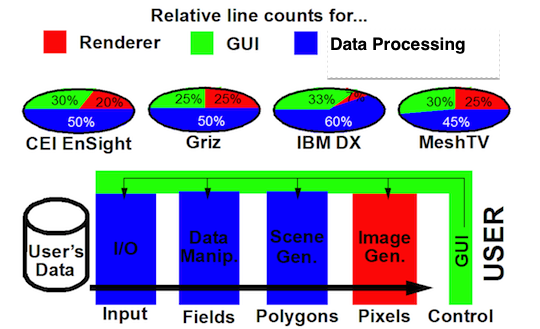
indicated that even back then 50% or more of the code in these tools was devoted to data processing aspects of the workflow. Since that time, it is almost certainly the case that this trend as continued to skew ever more towards data processing code development. For VisIt, the skew towards data processing code is closer to 80% for that and 20% for gui/rendering. As visualization tool developers, today we spend far less time now worrying about rendering or GUIs (though no one would argue that we could probably use better GUIs than we currently have) and spend more and more time developing data analysis algorithms and tools. In Visit, this kind of activity takes the form of adding new Operators, new Expressions, new Queries and new Python scripts that use these features of VisIt in batch processing workflows. Even the addition of new Plots to VisIt involves a combination of pre-rendering scene generation code development and data processing code development. These aspects of VisIt represent the kinds of activities that are good candidates for replacement by Big Data workflows.
So, here, we would like to see what’s involved in developing Spark enabled versions of some of VisIt’s common data processing operations. We investigated some Queries, some Expressions and some Plots (or equivalently Operators depending on how one differentiates the two in VisIt).
The basic data object in Apache Spark is a Resilient Distributed Dataset or RDD. The RDD interface is documented here. Spark provides a variety of useful operations on RDDs including and, in particular, for RDDs composed of key-value pairs. Apache Spark’s execution model is quite simple. A python script called a driver runs first and then submits copies of itself (and whatever other related Python and Java scripts may be needed) to a number of instances of itself running on the cores of a cluster. Now, there is a lot more to it than that. In particular, Spark includes a sort of dataflow paradigm for how a chain of RDD operations can be knitted together and how to manage caching and persistence of intermediate RDDs as well as how to handle common data exchange scenarios between the driver and the followers. But, it’s still very simple to learn and use.
In Apache Spark, the key interface for creation of an RDD from data in persistent storage is one of either hadoopFile(), newAPIHadoopFile() or textFile(). A key aspect of these interfaces is that they enable the creation of a single Spark RDD from an arbitrary collection of file(s) specified via file globs.
This provides a very convenient mechanism for specifying query scope, that is the breadth of the database over which an operation is performed. Given the File and Directory Layout described above, a pseudo-globbing template template for initiating a query looks something like this. . .
from pyspark import SparkContext
from pyspark import SparkConf
sc = SparkContext()
rdd_data = sc.textFile("file://home/vdbroot/<user-name(s)>/<database-name(s)/<state-number(s)/<mesh-name(s)>/<block-number(s)/...)
For brevity, from now on we will not include the boilerplate python code. For example, the first step in performing an operation over all blocks of a specific mesh (say bar), of a specific timestep (say 0) of a specific database (say foo) of a specific user (say miller) would look something like…
coord_data = sc.textFile("file:/home/vdbroot/miller/foo/000000/bar/*/coords.txt.bz2")
whereas, the same operation over all timesteps would start like…
coord_data = sc.textFile("file:/home/vdbroot/miller/foo/*/bar/*/coords.txt.bz2")
The same operation over timesteps 0 through 150 would be
coord_data = sc.textFile("file:/home/vdbroot/miller/foo/000[0-1][0-5][0-9]/bar/*/coords.txt.bz2")
The wider the scope, the more compute resources (cores and memory) are required to complete the operation in a given time constraint.
In this section, we just describe the implementation of a NumNodes query. In VisIt, this query is intended to return the total number of nodes in the mesh and the number of nodes that are ghost nodes.
sc = SparkContext()
coord_data = sc.textFile("file:/vdbroot.silo/miller/%s/%s/%s/*/coords.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[3]))
all_node_count = coord_data.count() # .count() is a builtin RDD operaton
ghost_node_count = coord_data.filter(lambda line: line[19]=='1').count() #.filter is a builtin RDD operation
print "Number of nodes is", all_node_count
print "Number of ghost nodes is", ghost_node_count
The arguments to the script are the name of the database, the timestep of interest and the mesh of interest. The star character (‘*’) in the sc.textFile file glob tells it to perform the query over all blocks of the mesh.
Note that these operations are also possible directly through commands in the Unix shell. For example, if you cd to the appropriate sub-tree in the File and Directory hierarchy, these commands will perform the equivalent operation with Unix tools. In addition, these operations from the Unix shell are actually significantly faster than through Spark. But, this has to do with the fact that Spark is designed for really big data and has large, fixed overheads for job startup and completion.
# all-nodes count
% find . -name coords.txt.bz2 -exec bzcat {} \; | wc -l
# ghost-nodes count
% find . -name coords.txt.bz2 -exec bzcat {} \; | grep '0[a-zA-Z0-9#%]*,1,' | wc -l
We can make a small modification to the NumNodes query and have it return values on a per-block basis.
sc = SparkContext()
coord_data = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/%s/%s/*/coords.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[3]))
# the "block-id" field of a node's key is characters in the slice [8:12]
nodes_per_block = coord_data.map(lambda line: line[8:12]).countByValue()
# the indication of a whether a node is ghost or not can be found by a '1' or '0' in char 19
ghost_nodes_per_block = coord_data.filter(lambda line: line[19]=='1').map(lambda line: line[8:12]).countByValue()
for p in sorted(nodes_per_block):
print "Block %d has %d nodes and %d ghost nodes"%(AsciiKeyToIndex(p),nodes_per_block[p],ghost_nodes_per_block[p])
Both of the above queries involved questions about the nodes only. So, we read only the coords.txt.bz2 files to satisfy the query.
We define functions min() and max() that perform a reduction of two RDD values into a new value and then pass these functions to the RDD’s reduce() operator. This approach involves two passes over the RDD, one for the min value and another for the max value.
def min(a,b):
if a[1] <= b[1]:
return a
return b
def max(a,b):
if a[1] >= b[1]:
return a
return b
sc = SparkContext()
# each textual line is read and a KV RDD is created with mesh entity key as Key and the variable's value as Value
var_kv = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/%s/*/%s.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[3]))\
.map(lambda line: (line[0:18], float(line[19:])))
min_kv = var_kv.reduce(min)
max_kv = var_kv.reduce(max)
print "Minimum value at %s is %g\n"%min_kv
print "Maximum value at %s is %g\n"%max_kv
We could do the same in a single pass with an operator that does a little more work as follows. . .
# constant indices for min value and max value in a min,max tuple
min = 0
max = 1
# Input is (key, (min,max))
def minmax(a,b):
if a[1][min] <= b[1][min]:
outmin = a[1][min]
else:
outmin = b[1][min]
if a[1][max] >= b[1][max]:
outmax = a[1][max]
else:
outmax = b[1][max]
return (a[0], (outmin,outmax))
sc = SparkContext()
# return a KV RDD with mesh entity as key and Value as (value, value) tuple (e.g. the variable's value on the entity duplicated)
var_kv = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/%s/*/%s.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[3]))\
.map(lambda line: (line[0:18], (float(line[19:]),float(line[19:]))))
minmax_kv = var_kv.reduce(minmax)
print "Minimum value is %g"%minmax_kv[1][min]
print "Maximum value is %g"%minmax_kv[1][max]
The above py-script has the advantage that it now performs only a single reduce() operation on the RDD. However, it has the disadvantage that it must create an RDD of a larger size; the values in the starting RDD is a tuple of min/max values that is initialized with the same value for each entity as both the min and the max. So, we may wind up doubling the memory usage to reduce the number of passes over the RDD we make. On the other hand, we really haven’t investigated how Spark actually performs the computation. It may expand individual RDD datums only one at a time. There may be an even better way to achieve memory usage of first approach with number of passes of second approach by using the actions aspects of RDDs more intelligently. We didn’t look into this yet.
Finally, it’s worth also noting that the same operations can be performed using Unix shell level commands
- find min using standard sort reversed (or could use ‘head’ rather than ‘tail’)
% find foo -name v.txt.bz2 -exec bzcat {} \; | sort -n -k 2 -t’,’ -r | tail -n 1- find max using standard sort
% find foo -name v.txt.bz2 -exec bzcat {} \; | sort -n -k 2 -t’,’ | tail -n 1
And, again, as was the case for the NumNodes query, the Unix implementation is much faster than the Spark implementation for the same reasons of fixed overheads to start and finish a job.
Here, we consider how to implement a simple binary expression; the plus (+) operator using a Spark RDD.
sc = SparkContext()
var1_data = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/%s/*/%s.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[3]))
nblocks = var1_data.map(lambda line: line[8:12]).distinct().count()
var1_kv = var1_data.map(lambda line: (line[0:18], float(line[19:])))
var2_kv = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/%s/*/%s.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[4])).map(lambda line: (line[0:18], float(line[19:])))
# forms a single kv-rdd with values from both variables where same node/zone appears
# as key of multiple pairs (one from var1 and the other from var2)
var12_kv = var1_kv.union(var2_kv)
# aggregates all values with same key by summing the value
# Here, you could replace '+' operator with any other of the operators in VisIt's binary
# operators and achieve the equivalent outputs
sum = var12_kv.reduceByKey(lambda a,b: a+b).partitionBy(nblocks,ParitionByBlockKeyField)
# This will save the partitioned data, block by block to HDFS (by default)
sum.saveAsTextFile("Operator+%s_%s.txt"%(sys.argv[3],sys.argv[4]))
In this operator, for the first time we are dealing with more than a single RDD. We read the first variable into an RDD and then immediately compute a count of distinct Block Ids (mesh entity key character slice [8:12]). Our aim is to use this count as a hint to Spark about how to partition the data when it writes it back out. We then form two key-value datasets of the form (Mesh Entity Key, Variable Value) and then union these. The union operation is not the same thing as a set-theoretic union. A better name for this Spark RDD operation would be maybe merge or combine. The fact is, after the union, the dataset may have multiple entries with the same key. In fact, we are depending on that in order for our summation here to work. The summation is performed using the reduceByKey operation on the RDD with a lambda of the form (a,b: a+b). That produces a sum result RDD where each mesh entity key appears only once in the RDD and the value is the sum of all entries in the original RDD with the same key. There may be better ways of implementing this operation. But, it was the first that occurred to me. The final action is to save the resulting sum RDD as a text file. But, we want the output text file to be structurally equivalent to the inputs. So, we use the partitionBy() method to ensure the dataset is partitioned according to Block Id field of the keys. Then, the saveAsTextFile saves this RDD, one file for each partition, to HDFS.
A recenter expression is even more interesting as it requires knowledge of the topology of the mesh. The goal is to average a nodal value to the zone-centers or a zonal value to the nodes. Here, we have implemented only the nodal-to-zonal recenter operation. We could easily add logic here to inspect the variable(s) specified and on the basis of mesh entity keys associated with the variable determine a variable’s centering. We read the topology.txt.bz2 data. Remember, that data is keyed by zonal mesh entities and the value is a tuple of nodal mesh entity references that comprise the zone. From the topology, we need to create a KV RDD that is either (nodeKey, zoneKey) or (zoneKey, nodeKey). For a nodal-to-zonal recenter, we need it to be keyed (nodeKey, zoneKey). And, that is what we’ve implemented here. However, doing the other direction is easy. We just adjust the lambda expression passed to the first .map() operator after textFile() reads the data.
sc = SparkContext()
zones = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/%s/*/topology.txt.bz2"%(sys.argv[1],sys.argv[2]))\
.map(lambda line: (line[0:18], (line[19:].split(',')[3:])))\
.flatMapValues(lambda x: x).map(lambda x: (x[1],x[0]))
var_data = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/%s/*/%s.txt.bz2"%(sys.argv[1],sys.argv[2],sys.argv[3]))
nblocks = var_data.map(lambda line: line[8:12]).distinct().count()
var_kv = var_data.map(lambda line: (line[0:18], float(line[19:])))
zones_with_vals = zones.join(var_kv)\
.values()\
.combineByKey(lambda value: (value, 1),\
lambda x, value: (x[0] + value, x[1] + 1),\
lambda x, y: (x[0] + y[0], x[1] + y[1]))\
.map(lambda (label, (value_sum, count)): (label, value_sum / count))\
.partitionBy(nblocks,ParitionByBlockKeyField)
zones_with_vals.saveAsTextFile("Recenter_%s.txt"%sys.argv[3])
The .textFile() operation reads the topology.txt.bz2 file producing an RDD consisting of lines of text of the form
“0000000000000005Fz,0,10,4,000000000000000l7M,000000000000000l7U,000000000000000l80,000000000000000l88”
This is a single line of a topology.txt.bz2 file. The first entry in a line is the zone key. The second entry indicates if the zone is a ghost zone or not. In this it example, it is not a ghost. The next entry, 10 here, indicates the zone type (using VTK’s zone type enumeration). A ‘10’ here indicates the zone is a tetrahedron. The next entry indicates that there are 4 node references for this zone. The remaining entries are the node keys for the nodes of this zone. Immediately after the
textFile() operator, the RDD has this form. But, a .map() operation is applied to it. The .map() operator takes a lambda expression that decomposes this line of text into a key and value pair, producing an RDD of the following form…
(0000000000000005Fz,[000000000000000l7M,000000000000000l7U,000000000000000l80,000000000000000l88])
This is a Python tuple, the first value of which is a zone key and the second value of which is a python list of node keys. Note that a basic constraint for a
.map() operation is that it produces one output entry for each input entry. However, a .flatMapValues() operation can produce multiple output entries for a single input. So, we use a .flatMapValues() operation to change the RDD to the form
(0000000000000005Fz,000000000000000l7M)
(0000000000000005Fz,000000000000000l7U)
(0000000000000005Fz,000000000000000l80)
(0000000000000005Fz,000000000000000l88)
where a zone with N node references results in N KV pairs in the output dataset. The zone key is duplicated several times, once for each zone/node pair. However, the final
.map() after reading the topology data reverses the order of keys and values in the tuple, producing a (nodeKey, zoneKey) RDD of the form
(000000000000000l7M,0000000000000005Fz)
(000000000000000l7U,0000000000000005Fz)
(000000000000000l80,0000000000000005Fz)
(000000000000000l88,0000000000000005Fz)
Next, we read the nodal variable data producing a KV RDD of the form
(000000000000000I7M,0.092)
(000000000000000I7U,0.097)
(000000000000000I80,0.093)
(000000000000000I88,0.090)
Where the KV pairs are (nodeKey, variable value) pairs.
Next, we .join() these two RDD’s together. The join operation produces an output value for each key that appears in both inputs. The resulting output is a KV pair where the key is the common key and the value is a python list combining the values from all the common keys. So after the .join() operation we have something like…
(000000000000000l7M,[0000000000000005Fz,0.092])
(000000000000000l7U,[0000000000000005Fz,0.097])
(000000000000000l80,[0000000000000005Fz,0.039])
(000000000000000l88,[0000000000000005Fz,0.090])
At this point, we really do not need the key part of this RDD anymore. We need only the value part which is itself another KV pair. So, we use
.values() operator to strip of the node keys leaving an RDD of the form
(0000000000000005Fz,0.092)
(0000000000000005Fz,0.097)
(0000000000000005Fz,0.039)
(0000000000000005Fz,0.090)
where the keys are zoneKeys and the values are the nodal variable’s values that contribute to the zone. So, the final step is to perform a
.combineByKey() operation in which we sum all the values associated with each key and then divide that sum by their count (e.g. average). Then, we save the resulting zone key’d RDD back to the database.
Here we consider what’s involved to find all zones containing 3 or more materials. What is interesting is that we can support this query over the entire Spark database. That is all users, databases, timesteps, meshes and blocks. What’s further amazing is how many lines of code we need to write to implement it, 2 plus the 7 lines for the count_mats() method.
def count_mats(volfracs):
nmats = 0
for i in range(len(volfracs)):
if (volfracs[i] > 0):
nmats += 1
return nmats
sc = SparkContext()
material_data = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/*/*/*/materials.txt.bz2"%(sys.argv[1]))
material_counts = material_data.map(lambda line: (line[0:18], count_mats([float(v) for v in line[19:].split(',')]))).filter(lambda p: p[1]>2).collect()
The first line reads the material data producing an RDD of lines from the materials.txt.bz2 files
0000n004000h0002gv,0,0.882,0.118
0000n004000h0002gD,0,1,0
0000n004000h0002gL,0,1,0
0000n004000h0002gT,0,0.711,0.289
0000n004000h0002g#,0,0.721,0.279
0000n004000h0002h7,0,0.913,0.087
This is a 3 material problem. Some of the lines above indicate the associated zone is clean in that material (volume fraction entry is 1.0 for a given material) while others indicate there is mixing. The second line produces a KV RDD of the form (zoneKey, # materials) which is the
.filter()‘d to include only those entries where the # materials > 2. We then .collect() all these back to the python driver. At this point, we have a list of all the zoneKeys with 3 or more materials as well as the actual count of the materials. To print them out in a nice way, we have code that decomposes the zone keys into sub-fields and the uses those and indices into their respective indexing files (users.txt.bz2 or dbs.txt.bz2 or states.txt.bz2, etc.) to produce useful output.
for zone in material_counts:
userIdx = zone0[0:2]
f = os.popen(“bzcat /home/training/vdbroot.silo/users.txt.bz2”,“r”).readlines()
for ul in f:
u = ul.rstrip().split(’,‘)
if userIdx == u0:
userStr = u1
dbIdx = zone0[2:5]
f = os.popen(“bzcat /home/training/vdbroot.silo/%s/dbs.txt.bz2”%userStr,“r”).readlines()
for dbl in f:
db = dbl.rstrip().split(’,‘)
if dbIdx == db0[2:]:
dbStr = db1
stateIdx = zone0[5:7]
f = os.popen(“bzcat /home/training/vdbroot.silo/%s/%s/states.txt.bz2”%(userStr,dbStr),“r”).readlines()
for sl in f:
s = sl.rstrip().split(’,‘)
if stateIdx == s0[5:]:
stateStr = s1
meshIdx = zone0[7:8]
f = os.popen(“bzcat /home/training/vdbroot.silo/%s/%s/%s/meshes.txt.bz2”%(userStr,dbStr,stateStr),“r”).readlines()
for ml in f:
m = ml.rstrip().split(’,‘)
if meshIdx == m0[7:]:
meshStr = m1
blockIdx = zone0[8:12]
f = os.popen(“bzcat /home/training/vdbroot.silo/%s/%s/%s/%s/blocks.txt.bz2”%(userStr,dbStr,stateStr,meshStr),“r”).readlines()
for bl in f:
b = bl.rstrip().split(’,‘)
if blockIdx == b0[8:]:
blockStr = b1
zoneIdx = AsciiKeyToIndex(zone0[12:]) / 8
print “user=%s, db=%s, state=%s, mesh=%s, block=%s, zone=%d has %d materials”%(userStr,dbStr,stateStr,meshStr,blockStr,zoneIdx,zone1)
The most significant outcome of designing this query is that it is possible with just a few lines of code to invoke the query over the entire Spark database or any portion thereof. This is remarkable! Doing the query over time, or over different databases or for different users requires nothing special. Well that’s not quite true. As we widen the scope of the query to include all times and/or all databases and/or all users, the memory and compute resources necessary to service the query surely do increase. We haven’t had sufficient time to study the real scaling behavior yet. Nonetheless, from a software engineering standpoint, the ability to write and run sophisticated queries such as this with little or no effort is truly remarkable.
Here we consider what’s involved in creating a new kind of Plot in VisIt; a wire frame plot. That is a plot that displays only the extreme edges of a mesh. An example we produced is show below

The Spark code to produce the wire frame is. . .
sc = SparkContext()
coords = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/%s/%s/*/coords.txt.bz2"%\
(sys.argv[1],sys.argv[2],sys.argv[3]))\
.map(lambda line: (line[0:18],[float(x) for x in line[19:].split(',')[1:]]))
wireEdges = sc.textFile("file:/home/training/vdbroot.silo/miller86/%s/%s/%s/*/topology.txt.bz2"%\
(sys.argv[1],sys.argv[2],sys.argv[3]))\
.flatMap(lambda line: zoneEdges(line))\
.reduceByKey(lambda a,b: True)\
.filter(lambda pair: not pair[1])
wireNodes = wireEdges\
.flatMap(lambda pair: [(pair[0][0:18],1),(pair[0][18:],1)])\
.distinct()
wireCoords = coords.join(wireNodes).map(lambda x: (x[0], x[1][0]))
collectedEdges = wireEdges.collect()
collectedCoords = wireCoords.collect()
It turns out this code doesn’t work in general because it assumes an extreme edge is an edge that is not shared by any other zones. However, there are many meshes where extreme edges are those shared by 2 or more zones. A modification to the algorithm was designed but could not be implemented in the available time. In addition, the above code merely determines the wire frame nodes and edges. We also need to write those back out to properly formatted coords.txt.bz2 and toplogy.txt.bz2 files to be consistent with the rest of the database. The code to do that is a little more involved…
f = os.popen("bzip2 > WireFrame_%s_coords.txt.bz2"%sys.argv[3],"w")
for c in collectedCoords:
f.write("%s,0,%g,%g,%g\n"%(c[0],c[1][0],c[1][1],c[1][2]))
f.close()
f = os.popen("bzip2 > WireFrame_%s_topology.txt.bz2"%sys.argv[3],"w")
keyBase = AsciiKeyToIndex(sys.argv[4])
keyBase /= 8
keyBase += 1
i = 0
for e in collectedEdges:
# key,ghost,type,count,nodeKeys
f.write("%s,0,3,2,%s,%s\n"%(IndexToAsciiKey("",(keyBase+i)*8+1,len(sys.argv[4])),e[0][0:18],e[0][18:]))
i += 1
f.close()
We have demonstrated the ease with which Apache Spark can be applied to common data analysis and post-processing operations. Apache Spark is under active development by a large, commercial community of users. So, using it gives us the ability to leverage activities from commercial industry. In addition, we can perform operations on HPC data with variable scope, from just one block of one mesh of one timestep of one database of one user to many blocks of many meshes of many timesteps of many databases of many users. This flexibility alone is a huge attraction of Apache Spark. In addition, the code development expertise to use Apache Spark doesn’t require a lot of experience with MPI and/or threaded parallelism or other potentially exotic HPC technologies.
Writing algorithms that operate based on Spark RDD operations and KV pairs is at first somewhat foreign and difficult to wrap ones head around. But, after an initial period of confusion and discovery, it becomes very straightforward and productive.
The downsides to this approach for data analysis are the inefficiencies in storage and with those, inefficiencies in computation due to added data movement load. We believe persistent storage inefficiencies can be addressed. They aren’t too bad to begin with and with modest effort, they can be improved significantly. The bigger issues are the memory and data movement during actual processing of data. Python is a convenient way to write Spark applications but a single integer in python, for example, requires 24 bytes of storage. That’s a 6x storage inefficiency. For small strings, such as our mesh entity keys, the inefficiencies are much higher; maybe >200 bytes for an 18 character key string. It’s conceivable that either Java or Scala would be a better long-term language to use for real production computing environment’s using Apache Spark. This issue needs to be evaluated.
Simplicity in data model and simplicity in execution model are big reasons that it’s easy to be productive in Apache Spark with minimal training and expertise. At the same time, this simplicity is what leads to the storage inefficiencies. Additionally, some of the conveniences we’ve demonstrated in this tiny project can be attributed more to the uniformity that was imposed in the initial database design than on anything inherintly provided by Spark. In real life HPC computing centers, users don’t all use a common file system for storing databases, or a common way of organizing them in a directory hierarchy or a common file format for storing them. This aspect of the project overly simplifies these issues. On the other hand, it might also suggest some changes in the way HPC computing centers permit their users to store data that could be considered in future storage system designs.
There are important advantages to programmer productivity and database flexibility Apache Spark brings to the table. The resulting increases in programmer productivity may more than offset costs that may result from added storage or compute resources to support such an approach.
There are Big Data software stacks based on Apache Spark that are also considering the ability to run low-level operations on GPUs. A GPU-enabled Apache Spark stack would be a very interesting technology to consider for LLNL’s future data analysis and post-processing needs. It’s also possible that a C++ implementation of Spark would allow storage inefficiencies to be put to rest.
Finally, the reader has no doubt observed that this work has focused primarily on determining whether the operations are possible and less so on how well the operations can be performed. In other words, we have no useful time and space performance analysis of the algorithms we’ve written. We have outlined a number of the storage inefficiences in the persistent storage format and how those inefficiencies may impact run-time performance of various algorithms but have nonetheless neglected to study algorithm performance in detail. This is something that must be considered in future work.
Things to consider for future
- Addressing inefficiencies in persistent storage format used here
- Study time and space performance of algorithms developed using RDDs
- Investigae feasibility of extending the existing RDD interfaces to optimize key operations
- Learning how RDD partitioning can be controlled to affect performance
- Study of scaling performance of python scripts such as those produced here and methods in the RDD interface
- Time/Space performance advantages of either Java or Scala as compared to Python studied here
- Is there a NumPy enabled implementation of Spark where RDD’s are implemented using NumPy arrays? This would offer significant improvement in storage efficiency.
- The possibility of a C++ Spark-Lite that incorporates many of the same principles of Apache Spark but runs with less job startup overhead and lower memory inefficiencies