-
Notifications
You must be signed in to change notification settings - Fork 24
Silo_Hadoop_and_VisIt
Briefly,Hadoop is a scalable infrastructure for processing unstructured or semi-structured, often line-oriented, textual data. It is a key technology used by many commercial companies whose profits depend on the ability to quickly process very large volumes of textual data (e.g. Google searches of the entire internet). The basic technology was developed at Google and then later Hadoop was created as an open-source version of it.
The core technology of"Hadoop":http://hadoop.apache.org is a scalable, parallel operation called map-reduce_. Two operators, a mapper and a reducer) are distributed to some (possibly very large) number of nodes in a computing cluster. The mapper phase reads data from the Hadoop filesystem (hdfs), processes it in some way and emits data that is then passed on to the reducer phase. Typically, data is marshalled around in Hadoop as key-value pairs. Sophisticated workflows involve chains of map-reduce operations. A question many in HPC computing have been interested in is whether Hadoop and map-reduce technology can be gainfully applied to HPC problems. A possible area of leverage is in post-processing. In fact, a key capability of the VisIt visualization tool called "_DDFs":http://www.visitusers.org/index.php?title=DDF, maps very straightforwardly to Hadoop map-reduce operations.
On two different LLNL ShipIt days, I have investigated the application of Hadoop to processing mesh-based scientific data from Silo files. In my most recent attempt, I enlisted the help of Cyrus Harrison and Kathleen Biagas to test use of Hadoop in post-processing. In particular we focused on Hadoop’s streaming interface which admits the use of python (or any executable for that matter) to implement mappers and reducers. Hadoop’s streaming interface does not permit access or control of all of Hadoop’s internals (you need to use Java for that) but is a sufficient interface to investigate and prototype capability. We implemented a simple VisIt DDF with Hadoop. The basic approach we took is described next.
We defined a line-by-line text file format in which each line is a JSON text object defining one zone of a mesh including the coordinates and node ids of all the nodes of the zone, all zonal variables names and values defined, all nodal variable names and values and a unique key for the zone to help disambiguate the zone from all other zones in all other meshes in this and all other Silo files we may wish to include in HDFS now or in the future. An example of one line from the Silo file, globe.silo, is shown below.
{"pt_ids": [1, 12, 67, 56, 0, 11, 66, 55], "zonals": {"EQPS": 0.0}, "cz": 0.024319999851286411, "cy": -0.0090499999932944775, "cx": -0.0090499999932944775, "key": {"domain": 0, "mesh": "Mesh", "obase": "exballs", "zone": 0, "time": 0.0, "dbfile": "/g/g11/miller86/visit/trunk/data/exodus_test_data/balls.exodus", "cycle": 0}, "zone_type": 12, "pts": [{"nodals": {}, "point_id": 1, "coords": [-0.010300000198185444, -0.010300000198185444, 0.023040000349283218]}, {"nodals": {}, "point_id": 12, "coords": [-0.0078000002540647984, -0.010300000198185444, 0.023040000349283218]}, {"nodals": {}, "point_id": 67, "coords": [-0.007799999788403511, -0.007799999788403511, 0.023040000349283218]}, {"nodals": {}, "point_id": 56, "coords": [-0.010300000198185444, -0.007799999788403511, 0.023040000349283218]}, {"nodals": {}, "point_id": 0, "coords": [-0.010300000198185444, -0.010300000198185444, 0.025599999353289604]}, {"nodals": {}, "point_id": 11, "coords": [-0.0078000002540647984, -0.010300000198185444, 0.025599999353289604]}, {"nodals": {}, "point_id": 66, "coords": [-0.007799999788403511, -0.007799999788403511, 0.025599999353289604]}, {"nodals": {}, "point_id": 55, "coords": [-0.010300000198185444, -0.007799999788403511, 0.025599999353289604]}]}
Note that we did not bother to set up precision in printing floating point values so the floating point values contain 2-3x as many characters as truely necessary. In addition, nodes and nodal values that are shared between zones in the original silo file wind up being duplicated in this format. That is simply the nature of a zone-by-zone enumeration such as this. So, with the combination of ascii output, using too many characters in floating point values and duplicating data on shared mesh entities, the size of the resultant file in HDFS is 2-3 orders of magnitude larger than the original Silo file! We’ve investigated some solutions to this which are described in a later section. Rest assured, we believe it is possible to reduce this file size expansion problem to under a factor of 2-4x (thats times not orders of magnitude). By the way, HDFS automatically duplicates the file 3 times as part of its standard redundancy to protect against node failures.
First, we used Hadoop to do some very simple VisIt-like queries on the data such as to compute a NumNodes or NumZones query. Given the file format, a NumZones query is equivalent to wc -l on the resultant text file and indeed we confirmed the number of lines of the text file gives the same result as a NumZones query from VisIt on the equiv. Silo file.
A NumNodes query is a little different though. We wrote a python mapper, mapper.py
#!/usr/bin/env python
import sys
import jsonfor line in sys.stdin.readlines():
data = json.loads(line)
for p in data[“pts”]:
cr = [round(x,3) for x in p[“coords”]]
crs = [“%0.3f”%x for x in cr]
print “%s\t%d”%(str(crs),p[“point_id”])
This mapper reads each line of the Hadoopified Silo file, loads a json object (which is just a nested collection of python dictionary objects) for the line (zone) and then emits lines of the form
[‘-0.210’, ‘-0.047’, ‘-0.231’] 0
[‘-0.212’, ‘-0.047’, ‘-0.231’] 1
[‘-0.213’, ‘-0.050’, ‘-0.231’] 2
[‘-0.210’, ‘-0.050’, ‘-0.231’] 3
[‘-0.210’, ‘-0.047’, ‘-0.228’] 4
[‘-0.212’, ‘-0.047’, ‘-0.228’] 5
which are the node’s coordinates and its nodal id for each of the nodes defined for the zone. Note that for nodes shared between zones, those nodes will be emitted multiple times. We then wrote a reducer that reads these lines and then indepenently counts unique cooredinate tuples and unique global node ids.
#!/usr/bin/env python
import sys
import jsonunodes = {}
- input comes from STDIN
for line in sys.stdin.readlines():
- remove leading and trailing whitespace
line = line.strip()if coord not in unodes.keys(): unodes[coord] = [] if nodeid not in unodes[coord]: unodes[coord].append(nodeid)
- parse the input we got from mapper.py
coord, nodeid = line.split(‘\t’, 1)
nodeid = int(nodeid)for node in unodes.keys():
unodes[node].sort()
print ‘%s\t%s’% ( node, len(unodes[node]) )
which results in output lines like so…
[‘-0.197’, ‘0.057’, ‘-0.123’] 93503
[‘0.118’, ‘0.042’, ‘-0.112’] 129281
[‘0.209’, ‘-0.052’, ‘-0.094’] 238349
[‘0.150’, ‘-0.049’, ‘-0.211’] 376362
[‘0.045’, ‘0.054’, ‘-0.093’] [427069, 460685, 460688]
[‘0.210’, ‘0.032’, ‘0.003’] 283092
[‘0.058’, ‘-0.055’, ‘-0.231’] 74994
[‘0.206’, ‘0.003’, ‘-0.173’] 131678
[‘0.144’, ‘-0.062’, ‘-0.226’] 77144
[‘-0.208’, ‘-0.048’, ‘-0.107’] [207711, 207731, 284927, 284928]
[‘-0.048’, ‘0.019’, ‘-0.155’] 108410
[‘0.198’, ‘-0.037’, ‘-0.197’] 11088
[‘-0.055’, ‘-0.052’, ‘-0.092’] [296659, 296666]
where each unique coordinate tuple has associated with it all the unique global ids. Note that the above data is from a poorly defined mesh from VisIt’s test suite. It has ~470,000 nodes but over 100,000 are duplicated (e.g. have same geometric coordinates but different node ids). For the mesh from globe.silo, for example, each line would contain just a single node id and
wc -l on the resulting output text file will produce the NumNodes query result from VisIt.
We ran these simple map-reduce operations on VisIt’s NASTRAN test data waterjacket.nas on bigfoot like so…
#!/bin/sh
hdfs dfs -rm -r -f wjnas_mesh_unodes
hadoop jar /usr/lib/hadoop-0.20-mapreduce/contrib/streaming/hadoop-streaming-2.0.0-mr1-cdh4.2.0.jar \
-file ./unodes.py -mapper ./unodes.py \
-file ./unodes_reducer.py -reducer ./unodes_reducer.py \
-input wjnas_mesh_0.0_0000.txt \
-output wjnas_mesh_unodes
rm -f wjnas_mesh_unodes.out
hdfs dfs -getmerge wjnas_mesh_unodes wjnas_mesh_unodes.out
Of the 471016 nodes in the input mesh, only 365270 have unique coordinate values. It turns out this particular mesh data, which was chosen somewhat randomly, was probably a poor choice for testing due to both the node duplication we discovered as well as the fact that the input file was in ascii and coordinate values had only 3 decimal digits of precision. Not knowing this originally caused some problems for our ShipIt exercise and we abandon using this dataset for the remainder of the exercise. However, it did suggest an interesting problem to try to solve with Hadoop and that was to write an algorithm to remove duplicate nodes from a mesh. More on that later.
We next attempted to implement a VisIt DDF (Derived Data Function aka Equivalence Class Function). DDF’s are an excellent match for Hadoop because they essentially do not depend at all on the fact that the input data represents some kind of a mesh with interrelationships between various entities in the mesh. Instead, DDF’s treat a mesh more or less as a point cloud in a somewhat arbitrarily chosen, multi-dimensional feature space and then map entites from the mesh into that features space.
For this part of the project, we used noise.silo dataset, converted into HDFS using the approach described above. We defined the mapper as…
#!/usr/bin/env python
import sys
import json
import osddf = json.loads(os.getenv(“VISIT_DDF”))
for line in sys.stdin.readlines():
zonals = data[“zonals”] bins = [] for d in ddf[“domains”]: name = d[“name”] if name in zonals.keys(): if zonals[name] >= d[“min”] and zonals[name] <= d[“max”]: bins += [int((zonals[name] – d[“min”]) / (d[“max”] – d[“min”]) * d[“nbins”])] if len(bins) == len(ddf[“domains”]) and ddf[“codomain”] in zonals.keys(): print “%s\t%s” % (json.dumps(bins), str(zonals[ddf[“codomain”]]))
data = json.loads(line)
if not “zonals” in data.keys():
continue
This mapper utilizes the fact that enviornment variables can be passed to Hadoop Map-Reduce jobs. The
VISIT_DDF variable defines a JSON objectification of a VisIt DDFAttributes object. For example…
{"domains":[{"dim":0,“name”:“hardyglobal_zonal”,min,max,nbinsdim,“name”:“radial_zonal”,min,max,nbins],“codomain”:"shepardglobal_zonal"}
The mapper reads each zone from the input, determines if the zone falls within the bounds of the DDF and, in particular, which x-bin and y-bin (in this case we are computing a 2D DDF) and then emits the value for the zone’s codomain as specified in the DDF attributes. In our example, the DDF we were trying to construct had its zeroth dimension broken into 30 bins for values of
hardyglobal_zonal variable in the range [0,6] and its one-th dimension broken into 50 bins for the values of the radial_zonal variable in the range [0,40]. The codomain variable we used was shepardglobal_zonal.
We defined the reducer as
#!/usr/bin/env python
import sys
import os
import time
import jsonfrom datetime import datetime
from datetime import timedelta
import string
import pickle
import json
import timedef now():
return datetime.now()def tdump(s):
tmp = pickle.dumps(s)
tmp = string.replace(tmp,‘\n’,‘@’)
return tmpdef tload(s):
tmp = string.replace(s,‘@’,‘\n’)
return pickle.loads(tmp)def ms_delta(tstart,tstop):
dt = tstop – tstart
ms = (dt.days * 24 * 60 * 60 + dt.seconds) * 1000 + dt.microseconds / 1000.0
return mst0 = tload(os.getenv(“VISIT_DDF_TIME”))
#
- Output for each key (bin indices)
- count of values in this ‘bin’
- sum of values in this ‘bin’
- average of values in this ‘bin’
- min of values in this ‘bin’
- max of values in this ‘bin’
- last elapsed time this key was seen
- maps words to their counts
binvals = {}
- input comes from STDIN
for line in sys.stdin:- remove leading and trailing whitespace
line = line.strip()if binkey in binvals: binvals[binkey][“count”] = binvals[binkey][“count”] + 1 binvals[binkey][“sum”] = binvals[binkey][“sum”] + binval; binvals[binkey][“avg”] = binvals[binkey][“sum”] / binvals[binkey][“count”] if binvals[binkey][“min”] < binval: binvals[binkey][“min”] = binval if binvals[binkey][“max”] > binval: binvals[binkey][“max”] = binval binvals[binkey][“time”] = ms_delta(t0,now()) else: binvals[binkey] = {} binvals[binkey][“count”] = float(1) binvals[binkey][“min”] = float(binval) binvals[binkey][“max”] = float(binval) binvals[binkey][“sum”] = float(binval) binvals[binkey][“avg”] = float(binval) binvals[binkey][“time”] = ms_delta(t0,now())
- parse the input we got from mapper.py
binkey, binval = line.split(‘\t’, 1)
binval = float(binval)
- write the tuples to stdout
- Note: they are unsorted
for v in binvals.keys():
print ‘%s\t%s’% ( v, json.dumps(binvals[v]) )
Note that this reducer actually computes several of VisIt’s DDF codomain operations all in one fell swoop. Ordinarily, in VisIt, one would re-execute the DDF for each of the count, min, max, avg, and sum operators.
We ran the map-reduce job on bigfoot like so…
#!/bin/sh
DDF=‘{"domains":[{"dim":0,“name”:“hardyglobal_zonal”,min,max,nbinsdim,“name”:“radial_zonal”,min,max,nbins],“codomain”:"shepardglobal_zonal"}’
hdfs dfs -rm -r -f ddf
sleep 1
VISIT_DDF_TIME=$(python -s gettime.py)
hadoop jar /usr/lib/hadoop-0.20-mapreduce/contrib/streaming/hadoop-streaming-2.0.0-mr1-cdh4.2.0.jar \
-file ./ddf_mapper.py -mapper ./ddf_mapper.py \
-file ./ddfreducer.py -reducer ./ddfreducer.py \
-input noise_zonal_Mesh_0.0_0000.txt \
-output ddf \
-cmdenv VISIT_DDF=$DDF \
-cmdenv VISIT_DDF_TIME=$VISIT_DDF_TIME
rm -f ddf.out
hdfs dfs -getmerge ddf ddf.out
env PYTHONPATH=/g/g11/miller86/silo/trunk/install/lib python make_silo_from_hadoop_output.py $DDF foo.silo ddf.out
The last line of this script takes the textual output file produced by the map-reduce job and uses Silo’s python interface to create a Silo file representing the DDF’s mesh and variables (this resulted in uncovering a few bugs in Silo’s Python interface). We ran this on whatever number of nodes Hadoop decided to (I think it was 6) and then loaded this Silo file into VisIt to display the results. The results are provided below.


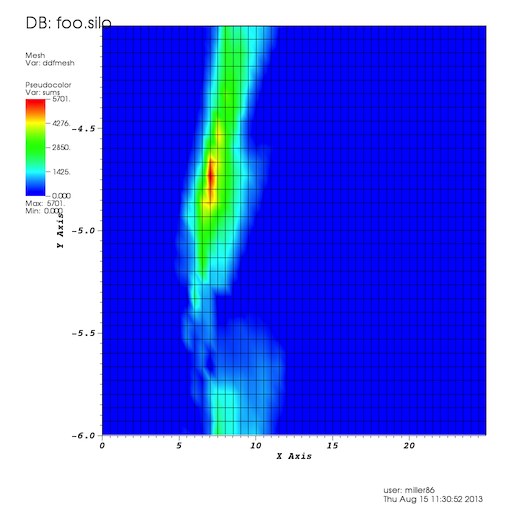

Unfortunately, the same DDF operations directly in VisIt did not yield similar results and we did not have time within the ShipIt day constraints to go back and figure out where we went wrong.
Nonetheless, the exercise did demonstrate the use of Hadoop in processor mesh-based scientific data.
First, thanks so much to Kathleen Biagas and Cyrus Harrison for jumping onto this project and providing so much help. It was my first experience doing a ShipIt exercise as a team and I really enjoyed it.
Also, I wanted to thank Jeff Long for helping out with last minute changes to our project and ensuring we had all the machine account access issues worked out.
Finally, a big thanks to Robin Goldstone for allowing us access to the Bigfoot Hadoop computing resource for performing our work. It is a really nice machine and a big improvment over my experiences with Hadoop on LLNL resources from the last ShipIt day.
Because we have limited time during ShipIt day exercise, there were aspects to running Hadoop Map-Reduce jobs that we didn’t have time to investigate further. Since ShipIt day, we have learned several useful things…
- Arbitrary (typically small) files can be passed to map-reduce jobs
- We could have passed DDF attributes using these files rather than as enviornment variables
- On LLNL’s Bigfoot at least, we can use C/C++ executables with Hadoop’s streaming interface
- I implemented the map and reduce operations in C++ and ran them on the same dataset as the python-based map and reduce operations and the C++ version ran 4x faster.
- I implemented the full map and reduce operation as one C++ executable and ran it on a single processor using the same HDFS file as input and it ran to completion in less time than 4-6 processor Hadoop map-reduce job in python.
- We can control the degree of parallelism Hadoop is using with these flags to Hadoop
-D mapred.reduce.tasks=32 \
-D mapred.map.tasks=32 \
However, the map.tasks count is only a suggestion to Hadoop. It may not obey that request. - You can exercise some degree of data-flow control between mappers and reducers controlling the order in which map outputs arrive at reducers as well as which map outputs arrive at which reducers
- This requires careful design of the key part of key-value pairs emmitted by the mappers as well as flags to Hadoop which control key partitioning and sorting
- We could reduce the size of our Hadoopified Silo files quite a bit. That is described in more detail below
- We need to consider using AMPLab’s Spark layer to remove latency and I/O associated with typical Hadoop streaming.
In our original design, we streamed the mesh into a text file where each line of the text file represented all of the data for a single zone of the mesh. We also did a poor job controlling the number of digits of decimal precision using in printing floating put values and introduce some other inefficiencies. This situation can be improved a lot by designing the hadoop-ified silo file a little differently.
The idea is to break the mesh up into tiny meshlets of a relatively small (but not necessarily constant) number of zones. Each meshlet would be output to a single line of the hadoop-ified file much like an ascii VTK file looks now. First, the nodes of the meshlet would be output. Then, the zones would be enumerated in terms of the meshlet’s tiny nodelist. Then, nodal and zonal variable data would be output. This would all be catentated into one, very long single line of the hadoop-ified file. By choosing meshlet size to be large enough to reduce duplication of mesh entities shared between meshlets, file size can be reduced. In addition, its concievable we could avoid the overhead associated with binary to ascii representation by using a binary representation for each meshlet. Finally, we can employ compression of each meshlet to reduce storage costs further. Taking these measures, I believe we could get the hadoop-ified file size down to 2-3x the size of the original Silo file.
During our ShipIt day exercise, we looked for some more interesting data than noise.silo and somewhat randomly choose a NASTRAN test dataset, waterjacket.nas. We then got mired in some oddities of this dataset giving us misleading results, given the assumptions we were making. We eventually discovered that this dataset has approximately 100,000 (25%) duplicate nodes (e.g. nodes at identical geometric positions but with different nodal ids). Additionally, the geometric coordinate data, which is ASCII, has only 3 decimal digits of precision. The dataset is shown below.

After ShipIt Day, it occurred to me to see if we could create a Hadoop Map-Reduce operator to identify and then remove duplicate nodes from this dataset. Additionally, I had reason to suspect that this dataset also included nodes with slightly different coordinates but that should really have been treated by the mesh as the same node. That is a bit of a harder problem to solve as it involves finding, for each node, any nodes that are so close in space as that they should be treated as the same node in the mesh, even if their cooridnates are slightly different.
A chain of map-reduce operations are involved. First, because we have need to compute distances fron one node to any other nodes, we need some kind of a spatial binning to avoid O(N^2) with the number of nodes, N. So, we need to compute spatial bounds and then bin the nodes up into different slabs, say in x-dimension. The first map-reduce operation does this; reading the original hadoop-ified silo data for waterjacket and accumulating spatial bounds as well as the smallest real edge length of any zone. Mappers emit this information as key-value paris like so…
xmin -0.214000
ymin -0.056000
zmin -0.214000
xmax -0.207000
ymax -0.026000
zmax -0.207000
emin 0.00100
A single reducer than reads all the mappers outputs and computes global bounds and the global minimum real edge length of any zone in the mesh. Note that any two nodes which are found closer together than this minimum edge length must really be the same logical node in the mesh even if their coordinates are slightly different.
Another map-reduce operation then takes the global bounds and minimum edge length as a data file that is passed to all mappers. The spatial bounds are then divided into say 10 bins in each of the x, y and z dimensions. Mappers read zones from the hadoop-ified silo file and then determine which spatial bin the zone’s nodes fall in. Nodes are emitted from the mappers with a part of their key indicating the spatial bin they fall in. Those nodes that are closer than emin to any spatial bin boundary are emitted multiple times, once for each spatial bin they are close to (note that this is somewhat equivalent to the notion of ghosting used in typical MPI-parallel based mesh algorithms). By setting up Hadoop’s partitioning to use the correct part of the key, we can ensure all the nodes that fall in the same spatial bin arrive at the same reducer. Each reducer could concievably be assigned more than one spatial bin to deal with. So, we next setup Hadoop’s shuffling algorithm to ensure all the nodes for one spatial bin arrive at a reducer before all the nodes for any other spatial bin. This way, each reducer need keep in memory at one time only those nodes for the current spatial bin on which it working. Without this part of the puzzle, each reducer would be required to read and then store up all nodes it will ever see before beginning work on any one spatial bin it may be assigned.
The reducer reads all the nodes for the current spatial bin (the indication that it has seen all the nodes for one bin will be the first occurence of a node for a different bin) and hashes them based on coordinates. Different nodes with same coordinates hash to the same place and those nodes are duplicate. Next, for each node in the current spatial bin, the reducer find all nodes also in that bin that are closer than emin in distance and stuffs those nodes into the same place in the hash. The result is that all nodes that are equal to or within emin of a given node are gathered together.
After running thise map-reduce operation for the waterjacket dataset, I discovered that there are no two nodes of different coordinate values that are closer than emin. So, all the duplicate nodes in the waterjacket mesh are identical duplicates. I have yet to take the final step and create a new mesh by removing duplicates from the existing one.
A problem with doing this in multiple map-reduce phases is that the input dataset is read multiple times. It would be interesting to see if we could get this all done in a single map-reduce step. Its concievable we could emit the global spatial bounds and minimum edge length information from each mapper multiple times but key’d for different reducers and shuffled so that it arrives at the reducers first ahead of all node data. Then, each reducer can be guaranteed of knowing the global spatial bounds and minimum edge length before processing any nodes.
Another thing I tought of was using some of the same stuff we started developing above to compute a sort of wireframe representation of a mesh. A simple thing is to find all nodes that are not share with other zones. That sort of gives the corners of a mesh and we experimented with that for some cases and it worked ok. Another thing to add to that is to find all edges that are shared only between two-zones (thats still a lot on the external surfaces though too) and, that connect corners and that have bound faces whose angle is large relative to faces around. them. This would be a very interesting algorithm to implement in Hadoop.