-
Notifications
You must be signed in to change notification settings - Fork 14
Detailed Description of Functionality
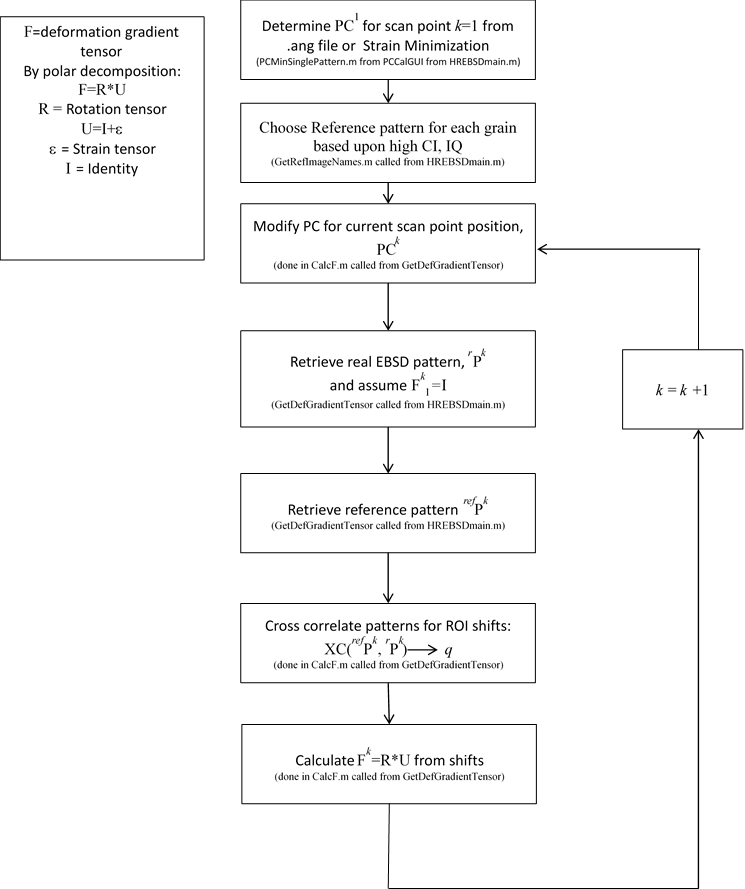
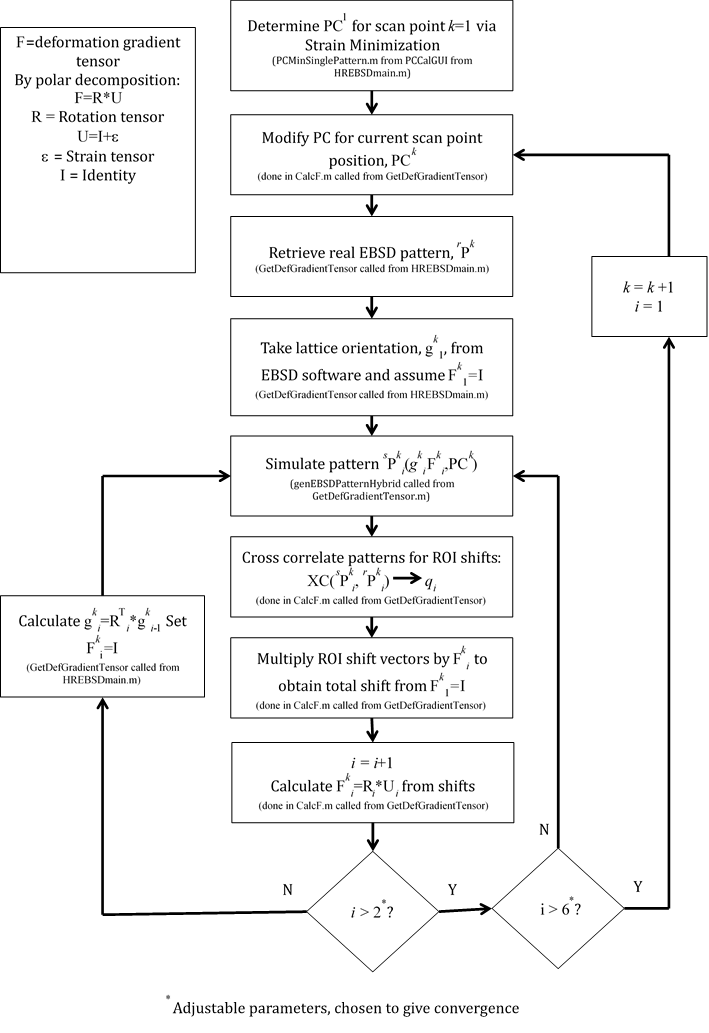
The basic function of this software is to use the cross-correlation method to determine the deformation gradient, F, between the crystal lattices for that of a reference EBSD pattern and that of a measured pattern [1-3]. The deformation gradient tensor, F, can then be split into a rotation and strain component: F=R*U where the strain tensor, ε, is given by U=I+ε. The reference pattern can either be a real pattern taken from a point on the sample (within the same grain as the measured pattern, since the orientations must be close), or a simulated pattern. The distortion between reference and measured pattern can also be thought of in terms of the elastic distortion tensor: =F-I. If the real pattern is taken close to the measured pattern then the elastic distortion between the points can be divided by the step size to provide the elastic distortion gradient, ij,k, which is fundamental to the continuum estimate of geometrically necessary dislocation (GND) content. Hence the following derived quantities can be extracted from the method:
*Relative strain (e.g. map of relative strain across a grain referenced from a chosen point in the grain) *GND density and Nye Tensor *GND content on different slip systems
*Absolute strain / tetragonality *Improved orientation measurement
###Detailed Program Architecture The main program is run from MainGUI.m. This sets up the main variables for the scan and then runs HREBSDMain.m. Various functions set up the necessary arrays (such as the list of image files), but the bulk of the work for the cross correlation method is done under GetDefGradientTensor.m and CalcF.m (which calls the actual cross correlation engine: custfftxc.m). These generate the F for each point, and thereby the strain maps. Along with these, the image simulation code is in genEBSDPatternHybrid.m, and the filtering is done in custimfilt.m. These files find the deformation gradients between reference and real patterns. In the case of simulated patterns, they also update the orientation for each scan point in line with the new calculated value. The returned Euler angles are stored in Settings.Data.phi1rn, PHIrn and phi2rn. The following two pages detail the main flow for the calculations of F for the real and simulated patterns. GND density is calculated in DislocationDensityCalculate.m which calls CalcF.m directly to determine the elastic distortion gradient between neighboring points in the two directions of the scan. From this it forms the available components of the Nye tensor, and an estimated bulk dislocation density.
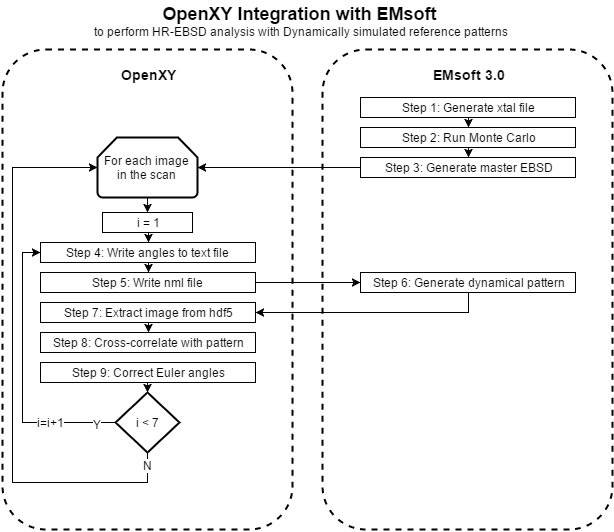
As described by its developers at Carnegie Mellon University, "EMsoft is an open source package for the simulation of electron microscopy imaging and diffraction modalities. It consists of a library with core routines for crystallography, symmetry, dynamical scattering, Monte Carlo simulations, and so on, plus a series of programs for the different modalities." OpenXY only uses EMsoft for the creation of the '.xtal' file, Monte Carlo simulation, Master pattern simulation and final EBSD pattern simulation. The diagram below depicts how OpenXY incorporates EMsoft. This [Installation Manual] (http://muri.materials.cmu.edu/wp-content/uploads/2015/11/EMsoft.pdf) gives a more in depth description of EMsoft's functionality.